Addressing the challenges of utilizing fresh frozen, FFPE and liquid biopsy for NGS
Investigating the roles of both DNA and RNA is essential in a range of scientific fields, including molecularbiology, genomics, biotechnology, and epidemiology. To do this, researchers need high quality nucleic acid samplesto meet the requirements for sensitive applications, such as next generation sequencing (NGS).
Using poor quality or contaminated samples for these applications might not yield accurate or biologically relevantresults. Consequently, there is a demand for reagents and techniques optimized for the isolation and purification ofhigh-quality DNA and RNA from a diverse range of biological sources.
These sources include fresh frozen tissue samples, formalin-fixed paraffin-embedded (FFPE) tissue blocks, and liquidbiopsy (e.g. a blood sample).
The isolation and purification of nucleic acids
DNA is generally stable when stored under suitable (frozen) conditions. It is also possible to conduct DNA isolation,purification, and storage from these different types of samples in batches. This is useful as it enables tissues tobe collected over an extended period of time, and the DNA isolated from them in a single day.
RNA is considerably less stable than DNA. Consequently, RNA isolation requires careful handling, processing, andstorage. If the smallest amount of RNase remains, it can degrade the sample.
Nucleic acid extraction and purification involves three key steps:
- Lysis: Breaking the cells open to expose the DNA and RNA.
- Lipid membrane removal: Treating the samples with a detergent such as Triton X-100 or NP-40.
- Nucleic acid precipitation: Achieved by adding alcohol to the sample.
There are also two optional steps during DNA isolation: removal of proteins in the sample(s) by adding a protease,for example proteinase K, and elimination of RNA with the use of an RNase, for example, RNAse A. These steps canminimize the impact of contaminants and increase DNA stability for long-term storage.
For RNA isolation, guanidium thiocyanate or a similar chaotropic agent isused to lyse the cells and denature RNase and DNase enzymes, protecting the sample(s). Once added, the samplesrequire robust shaking, usually vortexing, in the presence of a reducing agent, such as β-mercaptoethanol. Thisprocess breaks any disulphide bonds and inactivates any contaminant proteins present in the sample(s).
Importantly, and usefully for researchers, DNA and RNA can both be isolated from the same biological sample. Thisprocess involves extracting a total nucleic acid fraction and dividing it into two. One half is treated with DNase1, and the other RNase A.
Fresh frozen and formalin-fixed paraffin embedded samples in nucleic acidisolation
Fresh frozen tissue samples are widely considered the easiest type of tissue sample to process for nucleic acidisolation. The absence of chemicals and rapid freezing means that the nucleic acids, as well as the proteins withinthem, remain unmodified and in their native state(s).
This approach means fresh frozen tissue samples are well suited for mass spectrometry, Western blotting,next-generation sequencing (NGS), and quantitative real time PCR (qRTPCR), making them an invaluable resource formolecular biology and genetic studies.
There are many protocols available to process fresh frozen tissue samples, including manual methods developedin-house and a variety of commercial kits. These methodologies are rapid and straightforward, requiring thawing andmechanical disruption of the tissue samples in the presence of a chaotropic lysis buffer.
Another invaluable source of material, FFPE tissues, are held as large archives in pathology departments and clinicallaboratories around the world.
FFPE blocks enable the prolonged storage of clinical samples for investigation, preserving the tissue morphology forpathological examination, cancer diagnosis, and nucleic acids for isolation and downstream molecular analysis. Thesesamples are easy to collect, and of great interest to laboratories conducting clinical genomic studies,transcriptome analysis, biomarker identification, and chemo-resistance research.
The quality and integrity of the DNA and RNA from FFPE samples are affected by a number of factors (1,2):
- pH of the fixative.
- Length of tissue fixation.
- Age and storage condition of tissue blocks.
- Extraction method used.
Addressing the challenges with fresh frozen and FFPE samples
Despite the ease of using fresh frozen and FFPE samples, there are challenges. For fresh frozen, the most substantialchallenge is temperature. The samples rapidly deteriorate at room temperature. At the point of collection, thetissue needs to be frozen immediately as the longer it remains at room temperature, the more it degrades. Similarly,fresh frozen samples begin deteriorating when removed from storage, making rapid processing essential.
Controlling temperature from the moment of collection requires specialist equipment close to the site of surgery, sothat the sample can be frozen immediately after collection. Both short-term and long-term storage can also beexpensive, and there are risks of power outages and mechanical failures to mitigate, and simple human error with,for example, unsecured freezer doors that can affect sample quality.
While nucleic acid isolation from fresh frozen samples is uncomplicated, these challenges mean they are not collectedroutinely, and so biobanks typically have a smaller collection of frozen tissue specimens compared to FFPEspecimens.
The widespread availability of FFPE samples, and the importance of the information held within them, has drivensubstantial efforts to optimize FFPE protocols for extracting high-quality DNA and RNA. These efforts have led tothe development of several commercial extraction protocols for this specific purpose (3)
Nucleic acid extraction from fresh frozen and FFPE samples require different lysis buffers: sodium hydroxide-basedfor fresh frozen samples, and detergent and Tris-HCl-based buffer for FFPE samples (4).
Several commercial FFPE processing kits support paraffin removal using solubilization agents while other approachesdirectly melt the paraffin in tissue lysis buffer(s). Both approaches can minimize the loss of tissue during nucleicacid extraction.
However, nucleic acid extraction from FFPE samples is challenging even when using these commercial kits. Theprocesses typically yield inferior quality DNA and RNA compared to extraction from fresh frozen tissue samples. Thedetection of low frequency genetic variants from FFPE samples also poses considerable hurdles for researchers (5,6).
Formaldehyde, the active component of formalin, chemically cross-links the nucleic acids with the surroundingproteins and can also modify the DNA and RNA, creating artefacts. The formaldehyde deamination of cytosine bases todeoxyuracil is one such artefact, which can lead to a C-T conversion in later sequencing reactions.
Formalin can also damage and fragment the nucleic acids, and inhibit enzymes, including those used in reversetranscription and PCR amplification (7). This is of particular concern with NGS, as it is highly sensitive to the presence of contaminants or inhibitors.
The choice of the DNA and RNA extraction method used and the type of sample being processed appreciably impacts onnucleic acid integrity and downstream performance (8, 9, 10). These are particularly crucial for RNA due to itsinherent instability (11).
Liquid biopsy samples in nucleic acid isolation
Compared to surgery, which is necessary for obtaining tissues for freezing or formalin fixation, liquid biopsy isnovel, considerably less invasive, and can be sampled from a diverse range of fluids, including blood, urine,cerebrospinal fluid, saliva, stool, and lavage.
Alongside oncology, liquid biopsy has many clinical applications, including organ and transplant medicine andNon-Invasive Prenatal Testing (NIPT). The method has emerged due to substantial advances in the ability to isolatetotal cell-free DNA (cfDNA) and, within this, detect the circulating tumor DNA (ctDNA) or cell-free fetal DNA(cffDNA) species.
Unlike FFPE, liquid biopsy does not involve fixation or paraffin embedding, and does not have the temperaturechallenges noted when collecting, storing, or extracting nucleic acids from fresh frozen samples. Blood, forexample, is routinely collected and in the appropriate collection tubes, can be stored at room temperature forseveral days without a reduction in cfDNA quality. However, as sampling of blood for liquid biopsy is stillrelatively new, specialist biobanks are small and just beginning to be established, although there are a number ofgeneral serum-based repositories that can provide good insights albeit with sub optimal results.
Liquid biopsy and NGS
The advent of liquid biopsy is underpinned by NGS, though the NGS workflow can present its own sensitivitychallenges, particularly for liquid biopsy. These challenges include nucleic acid isolation, library preparation,and sequencing, the most critical being the ultra-low concentration of nucleic acids within these samples.
Unlike fresh frozen or FFPE samples, liquid biopsy contains only a small amount of cfDNA, and genomic DNAcontamination from lysed or apoptotic cells can interfere with detection. It is therefore essential to useefficient, highly sensitive technologies for the extraction and quantitation of nucleic acids from liquid biopsy forresearchers to obtain robust, reproducible data.
For example, incorporating liquid biopsy into recent oncology clinical trials has necessitated the development ofhigh‐throughput, reproducible, and efficient practices across the entire workflow, in particular, the nucleic acidextraction and library preparation steps.
These new methods and tools are now becoming widely available, with comparisons revealing that, in tandem withautomated systems, they can replace older, more laborious protocols. This is especially important whenhigh‐throughput cfDNA isolation is required (12).
Overcoming the hurdles of liquid biopsy nucleic acid isolation
Liquid biopsy samples contain a plethora of PCR inhibitors, including heparin and IgG (13, 14). As cfDNA is so scarce, a balance needs to be struckbetween obtaining high yield cfDNA and minimizing PCR inhibitors in any extraction strategy.
Also, although liquid biopsy has huge potential, the novelty of the approach compared to fresh frozen and FFPEapproaches means that there is limited clinical validation and standardization.
While the cfDNA in each liquid biopsy sample is extremely low, within this, the level of ctDNA is even lower andcommonly very fragmented (15). This fragmentation continues to present challenges during the DNA isolation processwhere the widespread loss of small size DNA fragments is common.
Unlike fresh frozen or FFPE samples, liquid biopsy also relies on ultrasensitive methods to isolate and detect smallquantities of nucleic acids. This can be misleading because it assumes that there is an unlimited amount of DNAavailable. If a liquid biopsy plasma sample contains one ctDNA molecule on average, not all samples will bepositive, even when using a method that can detect individual ctDNA molecules.
Irrespective of the type of sample, approaches for nucleic acid isolation from fresh frozen, FFPE, and liquid biopsyneed to be carefully optimized. Figure 1 shows a generalized overview of these approaches to DNA and RNA extractionfrom fresh frozen, FFPE, and liquid biopsy samples.
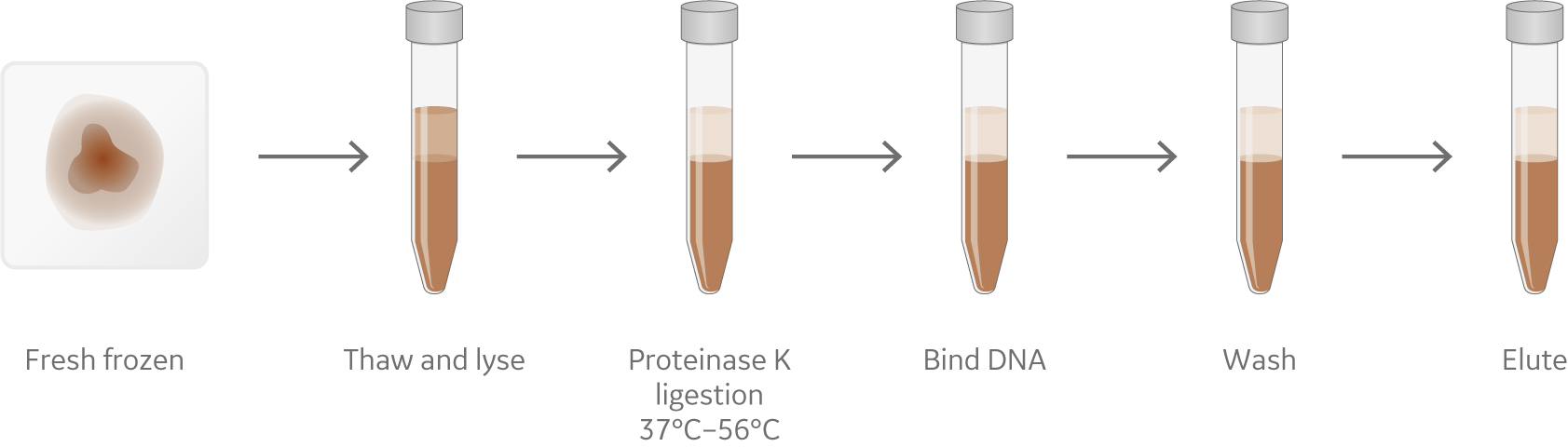
(A) Fresh Frozen tissues
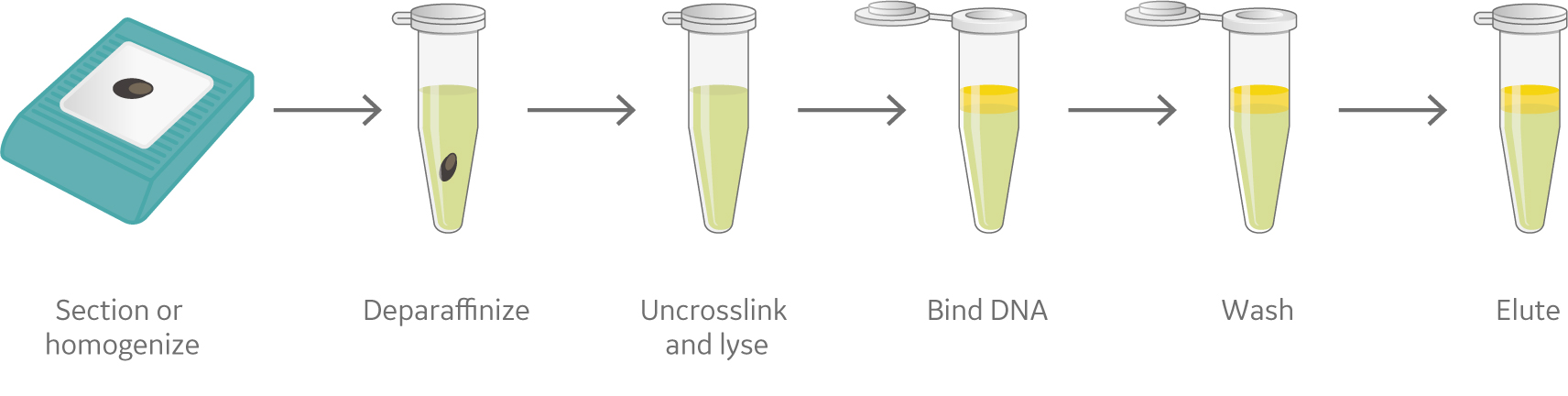
(B) Formalin-fixed paraffin-embedded tissue blocks
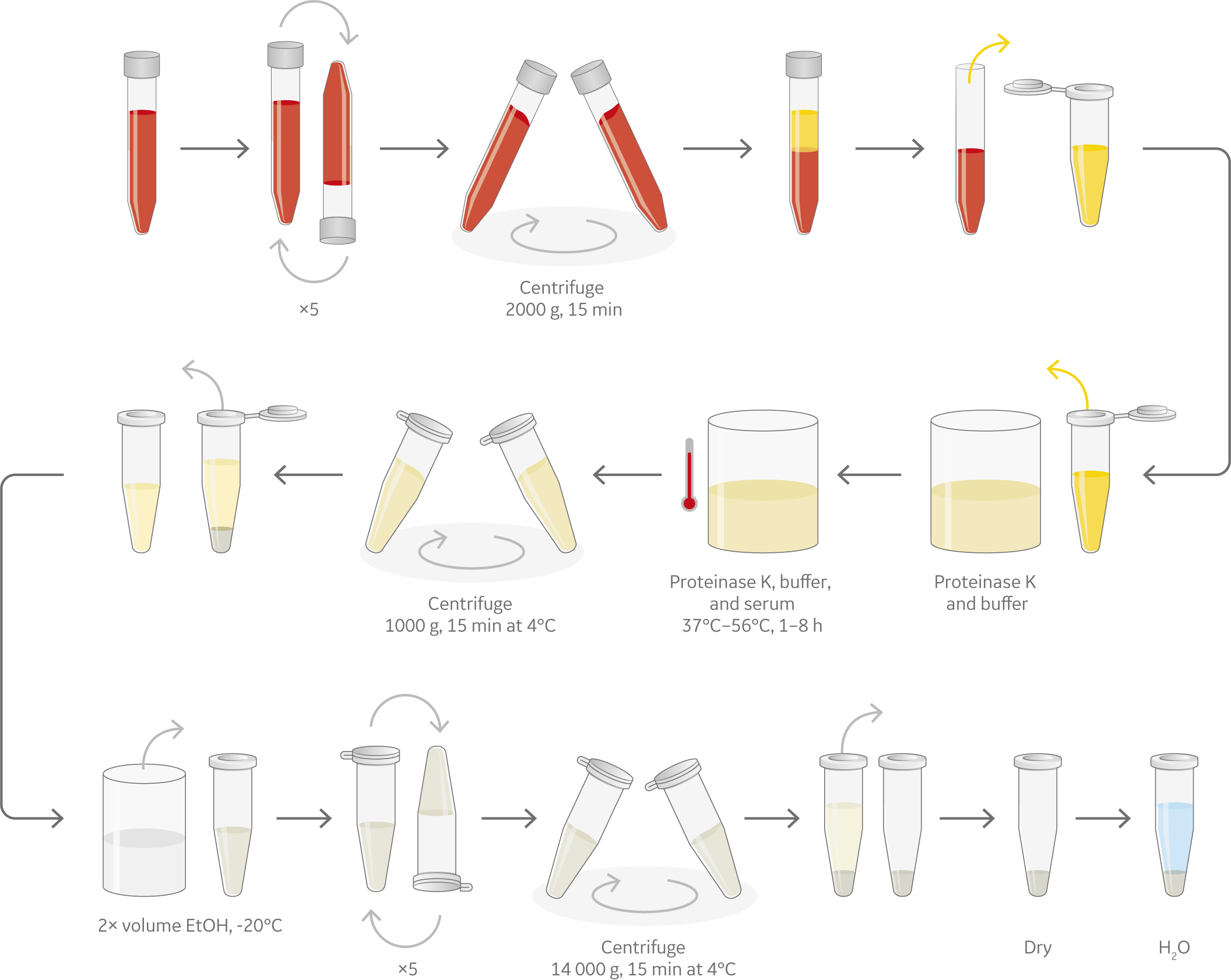
(C) Liquid biopsy
Fig 1 An overview of the nucleic acid isolation process (A) Fresh Frozen tissues, (B) Formalin-fixed paraffin-embedded tissue blocks and (C) Liquid biopsy.
Find out more about using and optimizing liquid biopsy workflows
Find out aboutSera-Xtracta Cell-Free DNA Kit for the extraction of cfDNA from plasma
Conducting NGS using extracted nucleic acids
NGS is a powerful platform that has enabled the sequencingof thousands to millions of DNA molecules simultaneously. In addition to genome sequencing, NGS has beenutilized for RNA sequencing (RNAseq) of tissue samples and even at the single cell level (scRNA-seq &snRNA-seq)and offers researchers the opportunity to characterize the transcriptome from fresh frozen, FFPE, andliquid biopsy samples with the greatest resolution.
RNA-seq can reveal the total cellular content of RNA including mRNA, rRNA, and tRNA, whether genes are turned on oroff in a cell, their level of expression, and their temporal regulation. These data can complement the genomicand protein data in matched fresh frozen, FFPE, or liquid biopsy.
NGS studies have demonstrated that, while nucleic acids isolated from FFPE tissues are more limited, with lowercoverage compared to matched fresh frozen tissue samples, these nucleic acids can pass the stringent NGS qualitycontrol requirements for sequencing if the integrity is good (16).
There is a need for consistent, reliable, and sensitive NGS reagents and workflows to tackle both these challengesand analyze the enormous volume of data generated from studying these complex diseases.
Find out how to avoid bottlenecks and delays in the NGSpipeline
Optimizing nucleic acid isolation from fresh frozen, FFPE, and liquidbiopsy
As long as storage and workflow temperature are closely monitored, processing fresh frozen tissue samples isrelatively straightforward. This is a practical challenge, overcome by careful planning and appropriateinfrastructure that enables straightforward sample collection and high-quality nucleic acid isolation. In contrast,FFPE and liquid biopsy samples need additional steps and care to ensure they generate reliable and reproducibledata.
Nucleic acid repair
An important recent development within the FFPE nucleic acid extraction workflow is the inclusion of nucleic acidrepair strategies designed to tackle artefacts introduced by the fixation and embedding conditions.
In these repair strategies, after heating to remove cross-links introduced by formalin, the DNA is accessible for thespecific removal of deaminated cytosine residues by the enzyme Uracil N-Glycosilase (UNG). This enzyme canspecifically remove artificially-induced uracils from the isolated nucleic acids and so reduce the likelihood ofinaccuracies in NGS output.
Nucleic acid amplification
Due to its low abundance, and despite the sensitivity of NGS, acquiring enough cfDNA by liquid biopsy for sequencingusually necessitates extracting large, sometimes variable, volumes (17). Some strategies suggest tackling this challengethrough amplification rather than increasing sample volume, for example, by Phi29 DNA polymerase-based rollingcircle amplification (RCA) of ligated cfDNA fragments. Alternatively, commercial library preparation kits employ lowcycle PCR using high-fidelity polymerases to minimize the risk of amplification bias.
Adding magnetism
In parallel to repair strategies, the incorporation of and improvements in magnetic beadtechnology have essentially removed centrifugation steps. This change has resulted in reduced sample lossduring nucleic acid isolation from fresh frozen, FFPE, and liquid biopsy samples and damage to the nucleic acids bymechanical force.
A magnetic bead-based approach also enables the researcher to isolate DNA based on a sizerange that can be tailored to suit the specific experimental requirements. Modulating this target size rangeis achieved by adding various ratios of magnetic beads to the sample(s) and supports NGS library and PCR fragmentclean-up, as well as dual size selection of libraries.
Surface chemistry options also enables researchers to use magnetic beads to specificallyisolate and extract mRNA for downstream applications, such as RNA-seq, real-time qPCR, microarrays,affinity purification, primer extension, and subtractive hybridization.
These improvements to the workflow enable the extraction and assessment of low input library preparations from anysample, but particularly FFPE and liquid biopsy. This would be a considerable challenge by any other method, andenables reliable whole exome sequencing and targeted panel sequencing applications for a greater range of samples.
The path forward for fresh frozen, FFPE, and liquid biopsy samples
While liquid biopsy is beginning to revolutionize oncology, there remains a clear need to analyze fresh frozen andFFPE samples in many medical fields. To meet this need, and obtain samples with the quality necessary for sensitivedownstream applications, deparaffinization, un-crosslinking, contaminant removal, nucleic acid degradation, purity,and library amplification all require consideration.
It is essential to carefully evaluate the approach used for nucleic acid isolation, irrespective of the startingbiological source. The methodology needs to be tested and validated beforehand to maximize the accuracy of thedownstream results obtained.
Each type of sample, fresh frozen, FFPE, and liquid biopsy, offers advantages but also challenges to the researcherfor nucleic acid isolation, summarized in Table 1.
Table 1. Summary table for the sources of nucleicacids available within various clinical biological sources, as well as the advantages and disadvantages
| Sample type | Description | Advantages and disadvantages |
|---|---|---|
| Fresh frozen tissue block | After surgical excision, tissues are snap-frozen in liquid nitrogen. These specimens are stored in liquid nitrogen or designated -150°C freezers before use. | Advantages
Disadvantages
|
| Formalin-fixed paraffin embedded (FFPE) block | Following surgical collection, tissue samples are fixed in formalin andparaffin-embedded. They are the most common source of archived material and stored at room temperature. | Advantages
Disadvantages
|
| Liquid biopsy | Aliquid biopsy is the sampling and analysis of non-solid tissue (e.g. blood). These samples are routinely collected and do not require immediate processing or storage. | Advantages
Disadvantages
|
FFPE-tailored reagents make the extraction of high-quality nucleic acids from FFPE samples achievable and enable theresearcher to conduct NGS with confidence. Older specimenscan be problematic due to variations in fixation or storage procedures. However, with an optimized approach takingaccount of these concerns, historic samples can still be suitable for many applications, including NGS and wholetranscriptome analysis.
Liquid biopsy approaches enable the isolation and detection of cfDNA and ctDNA, drawing on the strength of NGStechnologies now available to analyze ultra-low concentrations of nucleic acids. However, there is still nostandardized approach for liquid biopsy processing, and so there is a need to address genomic DNA contamination andPCR inhibitor presence for improved sensitivity.
The first step and challenge in the path forward for fresh frozen, FFPE, and liquid biopsy samples is reliablyisolating high-quality nucleic acids, after which generating a library, sequencing it, and analyzing the data arerelatively straightforward.
Find out more about improving the FFPE sample workflow or contact the ScientificSupport team to discuss any other aspects of the nucleic acid extraction workflow.
- Watanabe M. et al. Estimation of age-related DNA degradation from formalin-fixed and paraffin-embedded tissue according to the extraction methods. Exp Ther Med. Sep;14(3):2683-2688, (2017).
- Evers DL., et al. Paraffin embedding contributes to RNA aggregation, reduced RNA yield, and low RNA quality. J Mol Diagn. Nov;13(6):687-94, (2011).
- Kocjan BJ., et al. Commercially available kits for manual and automatic extraction of nucleic acids from formalin-fixed, paraffin-embedded (FFPE) tissues Acta Dermatovenerol Alp Pannonica Adriat. 2015;24(3):47-53, (2015).
- Wang JH., et al. DNA extraction from fresh-frozen and formalin-fixed, paraffin-embedded human brain tissue. Neurosci Bull. Oct;29(5):649-54 (2013).
- McDonough SJ., et al, Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PLoS One. Apr 11;14(4):e0211400, (2019).
- von Ahlfen S., et al. Determinants of RNA quality from FFPE samples. PLoS One. Dec 5;2(12):e1261, (2007).
- Masuda N., et al. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. Nov 15;27(22):4436-43, (1999).
- Heydt C., et al. Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics. PLoS One. Aug 8;9(8):e104566, (2014).
- Huijsmans CJ., et al. Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Notes. Sep 14;3:239, (2010).
- Janecka A., et al. Comparison of eight commercially available kits for DNA extraction from formalin-fixed paraffin-embedded tissues. Anal Biochem. May 1;476:8-10, (2015).
- Kresse SH., et al. Evaluation of commercial DNA and RNA extraction methods for high-throughput sequencing of FFPE samples. PLoS One. May 17;13(5):e0197456 (2018).
- van Dessel LF., et al, High-throughput isolation of circulating tumor DNA: a comparison of automated platforms. Mol Oncol. Feb;13(2):392-402, (2019).
- Schrader C., et al. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. Nov;113(5):1014-26, (2012).
- Sidstedt M., et al. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal Bioanal Chem. Apr;410(10):2569-2583, (2018).
- Jahr S., et al,DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res.Feb 15;61(4):1659-65, (2001).
- Bonnet E., et al. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PLoS One. Apr 5;13(4):e0195471, (2018).
- Gyanchandani R., et al. Whole genome amplification of cell-free DNA enables detection of circulating tumor DNA mutations from fingerstick capillary blood. Sci Rep. Nov 23;8(1):17313, (2018)