By Chike Ejiofor, Senior Field Application Scientist, Genomics and Diagnostics Solutions, Cytiva and Monika Seidel PhD, Senior Development Scientist, Genomics and Cellular Research, Cytiva
Summary
The newly developed Sera-Xtracta Virus/Pathogen Kit* meets the needs for:
- Simplified and rapid total nucleic acid purification chemistry
- For use with a variety of sample types (swabs, blood and samples in universal transport media)
- Removal of contaminants and inhibitors for the detection of low viral titer from 10 pfu/mL
- Consistent high yields with a simplified protocol that can be easily automated
Introduction
Infectious diseases affect millions of people every year. Particularly virulent and multi-drug resistant agents are increasingly responsible for infections with ever-expanding complexities. Molecular diagnostic laboratories and their test developers need to design, manufacture, and validate assays for these pathogenic agents. This requires the successful purification of high-quality nucleic acid which is essential to any molecular research and testing workflow. The nucleic acid purification process can be a bottleneck because sufficient nucleic acid from biological samples is required to meet a sensitivity threshold for an assay or tests which are designed to help mapping or making informed decisions on latent and active infections.
To address this bottleneck, Cytiva has developed a robust extraction chemistry optimized with SeraSil-Mag magnetic beads for total nucleic acid (DNA/RNA) from various sample types. Our optimized reagent chemistry ensures total nucleic acid is selectively bound to the superparamagnetic SeraSil-Mag bead while impurities are efficiently removed during the quick wash steps. The resulting high quality total nucleic acid is then eluted from the bead using suitable elution buffer of choice. The rapid workflow (Fig 1) gives pure and high-quality nucleic acid suitable for sensitive molecular testing including quantitative polymerase chain reaction (qPCR, RT-qPCR), droplet digital PCR (ddPCR), and next-generation sequencing (NGS).

Fig 1. Nucleic acid extraction workflow with SeraSil-Mag beads and buffer
This application note demonstrates the proof of concept for using our Sera-Xtracta Virus/Pathogen Kit for purification of DNA/RNA viruses and bacteria from oropharyngeal swabs and blood.Detection of Influenza A and SARS-CoV-2 from different sample types*
Following the kits protocol, the extraction efficiency of Sera-Xtracta Virus/Pathogen Kit was investigated using contrived oropharyngeal swabs samples, human clinical blood and murine blood samples as listed below:
- DNA/RNA viruses:
- Adenovirus (Type 14): Contrived clinical human blood sample (2008 clinical isolate)
- Influenza A (H3N2): Contrived clinical oropharyngeal swab (Copan Flocked swab UTM 306C) was spiked with 10^3 and 10^4 cp/mL of influenza A (H3N2) RNA sourced from a previous project (1)
- COVID-19: Contrived clinical oropharyngeal swab (Copan Flocked swab UTM 306C) was spiked with 10^3 and 10^4 cp/mL of COVID-19 RNA. The primers/probes are the N1 and RNase P assay recommended by CDC (2).
- Internal positive control for human nucleic acid was tested with RNase P
- Bacteria:
- Blood from 3 mice infected (challenge dose at 10^6 CFU/mL) with Mycobacterium tuberculosis (MTB)
- Oral Swab samples (BD Sterile Flocked swab. P/N 220528) from 2 consenting TB patients collected in 1.5 mL PrimeStore-MTM (Longhorn Vaccines and Diagnostics) and shipped at ambient temperature.
Using a simplified and rapid protocol total nucleic acid was extracted from each contrived sample (up to 400 µL sample volume) using Sera-Xtracta Virus/Pathogen Kit. Elution volume was set at 50 µL for all samples extracted.
Assay was set-up once with samples in duplicates, by adding 5 µL of eluted total nucleic acid to the mastermixes. Reverse transcription and qPCR runs were performed according to manufacturer’s user guide for the following kits:
- RT-qPCR for Influenza A (H3N2) and Covid-19: TaqPath™ 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific)
- qPCR for Adenovirus: (Type 14): Applied Biosystems™ TaqMan™ Fast Advanced Master Mix (Thermo Fisher Scientific)
- qPCR for MTB: PrimeMix MTB qPCR Blend (Longhorn Vaccines and Diagnostics)
Data in Figure 2 shows detection of Adenovirus Type 14 in human whole blood sample spiked with several dilutions. Detection of low viral titer at 10 pfu/mL indicates that the chemistry is suitable for sensitive applications with efficient recovery as reflected by the average Ct values across dilutions.
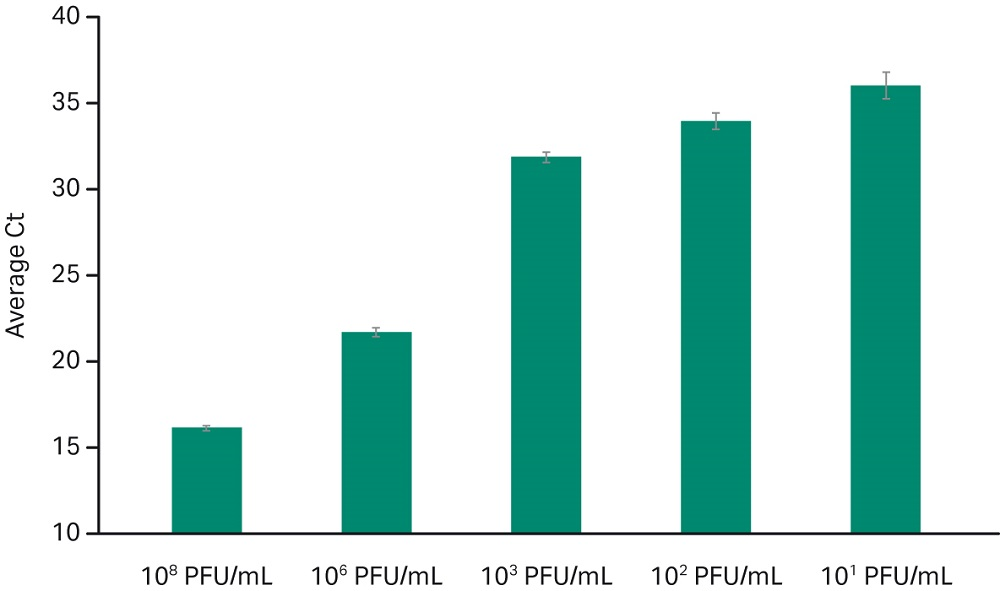
Fig 2. Adenovirus Type 14
The SeraSil-Mag formulation chemistry is robust enough to extract Mycobacterium tuberculosis DNA from both oropharyngeal swabs collected from tuberculosis patients and from blood samples collected from MTB challenged mice (Fig 3). These results show consistency across different patient blood samples and murine blood.
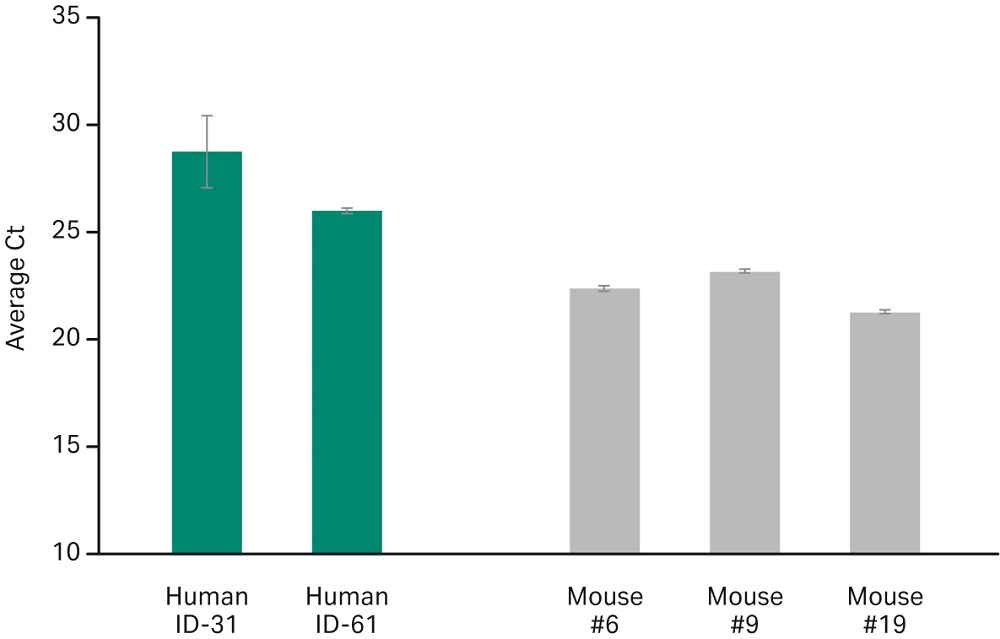
Fig 3. MTB Human and Murine blood
Due to the unstable nature of RNA viruses, their high surface and genome structure diversity make universal viral nucleic acid extraction challenging (3). With the onset of the COVID-19 pandemic caused by SARS-CoV-2 (RNA virus), a proof of concept was conducted to demonstrate that Sera-Xtracta Virus/Pathogen Kit is suitable for extracting viral RNA from oropharyngeal swabs. In this brief study, performance was compared to the RNAspin Mini Kit (Cytiva), a generic total RNA isolation kit. The data in Figure 4 shows both kits give excellent recovery and extraction efficiency from RNA viruses such as Influenza A (H3N2) and SARS-CoV-2. RNase P was tested to detect RNase P gene as an internal positive control for human nucleic acid.
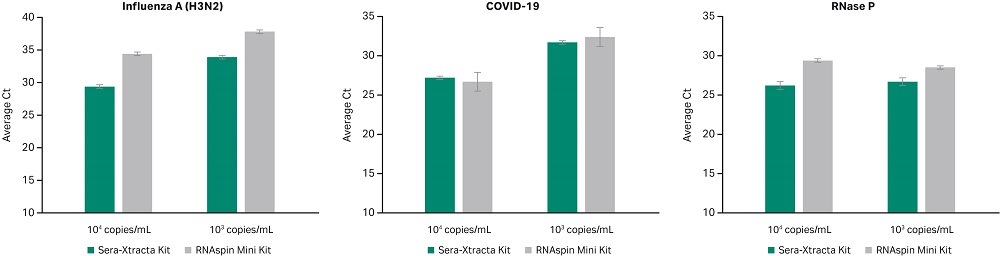
Fig 4. Detection of Influenza A and SARS-CoV-2 from RNA extracted using Sera-Xtracta Virus/Pathogen Kit and RNAspin Mini Kit
Recovery of synthetic SARS-CoV-2 RNA
Synesthetic viral RNA (SeraCare) covering nucleocapsid gene of SARS-CoV-2 was used to assess the performance and define the approximate limit of detection of the Sera-Xtracta Virus/Pathogen Kit in the experimental setting relevant to COVID-19 detection from human swab samples. Viral RNA was spiked at between 1 to 100 copies/µL into a diluent consisting of a suspension of human cells (~3x 10^5/mL) in 400 µL of PrimeStore MTM (FDA approved transport media, Longhorn Vaccines and Diagnostics) to mimic a clinical sample. Un-spiked cell suspension (no viral RNA) served as the assay control to eliminate the possibility of false positives. The samples were processed using Sera-Xtracta Virus/Pathogen Kit, eluted in 50 µL of nuclease-free water and subjected to Real-Time RT PCR assay following Centers for Disease Control and Prevention (CDC) protocol. Briefly, extracted samples (5 µL of eluant per well) were run in technical duplicates using TaqPath™1-Step RT-qPCR Master Mix, CG (ThermoFisher Scientific) and CDC 2019 Novel Coronavirus (2019-nCoV) Diagnostic Panel primers (N1, N2, N3 targeting three regions of Covid-19 nucleocapsid gene and RNase P primers targeting human RNase P gene).
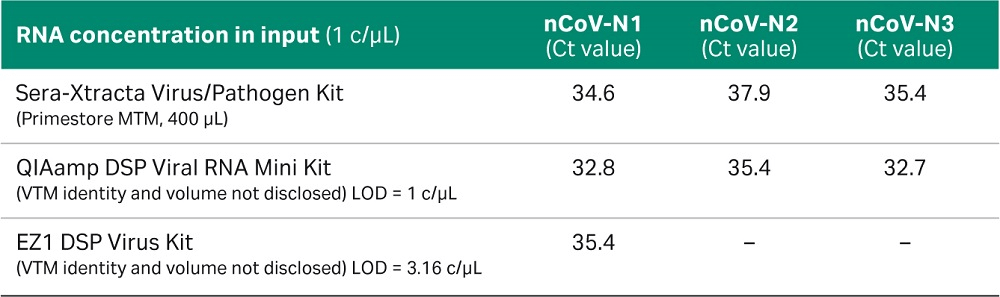
The results, presented in Figure 5, confirm that the Sera-Xtracta Virus/Pathogen Kit is robust and sensitive enough for reproducible detection of low viral titer down to 1c/µL in the input sample. For comparison with the CDC published data for the two referenced QIAGEN™ kits, please refer to the table below (CDC Division of Viral Diseases, CDC-006-00019).
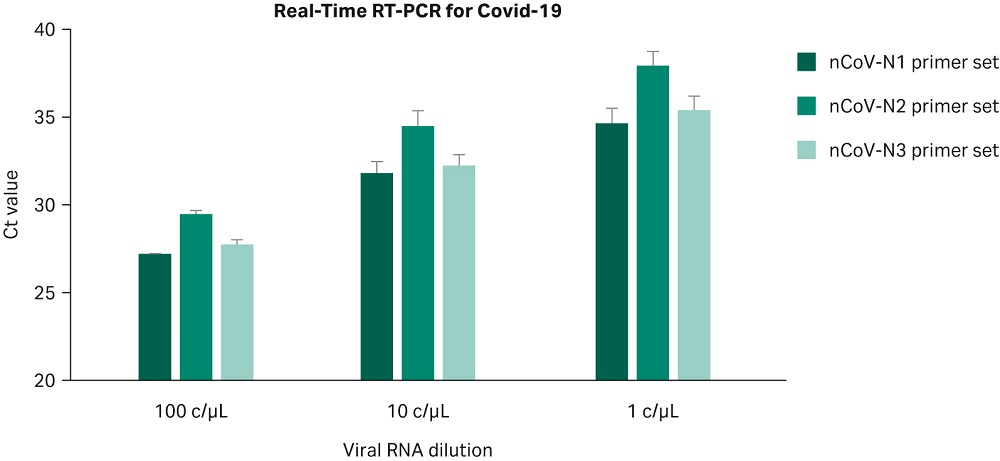
Fig 5. Ct values obtained for each of the three COVID-19 specific primer sets with varying amounts of the viral synthetic RNA in the input sample as described on the graph. Values averaged from four independent experiments with the exception for 100 copies/µL that was performed twice. Error bars represent standard deviation.
Table 1. Ct values obtained for each of the three COVID-19 specific primer sets at one copy /µL in reference to the values published by CDC Division of Viral Diseases, CDC-006-00019 for two listed QIAGEN kits. Please note that the transport media identity and input volume has not been disclosed in the referenced document.
Comparative performance of Sera-Xtracta Virus/Pathogen Kit
Synesthetic viral RNA (Microbiologics, Cat No 9009) covering nucleocapsid gene of SARS-CoV-2 was used to compare the performance of Sera-Xtracta Virus/Pathogen Kit with QIAamp™ MinElute Virus Spin Kit (QIAGEN). Although the QIAGEN kit is based on silica columns and involves the use of carrier RNA, thus differs considerably from Sera-Xtracta Virus/Pathogen Kit, it has been chosen as a benchmark due to the kit being recommended by CDC for the extraction of COVID-19 RNA. Viral RNA was spiked at 1, 100 and 10^4 copies/µL into a diluent consisting of a suspension of human cells (~3x 10^5/mL) in 200 µL of PrimeStore MTM to mimic a clinical sample. The samples were processed using Sera-Xtracta Virus/Pathogen Kit and QIAamp MinElute Virus Spin Kit following manufacturer’s protocol, eluted in 50 µL of nuclease-free water and subjected to Real-Time RT PCR assay following CDC protocol. Briefly, extracted samples (5 µL of eluant per well) were run in technical duplicates using TaqPath™1-Step RT-qPCR Master Mix, CG (ThermoFisher Scientific) and CDC 2019 Novel Coronavirus (2019-nCoV) Diagnostic Panel primers (N1, N2, N3 targeting three regions of Covid-19 nucleocapsid gene and RNase P primers targeting human RNase P gene).
Data from three independent experiments are presented in Figure 6 and confirm that both kits are able to detect the presence of viral RNA down to 1 c/µL in the input sample.
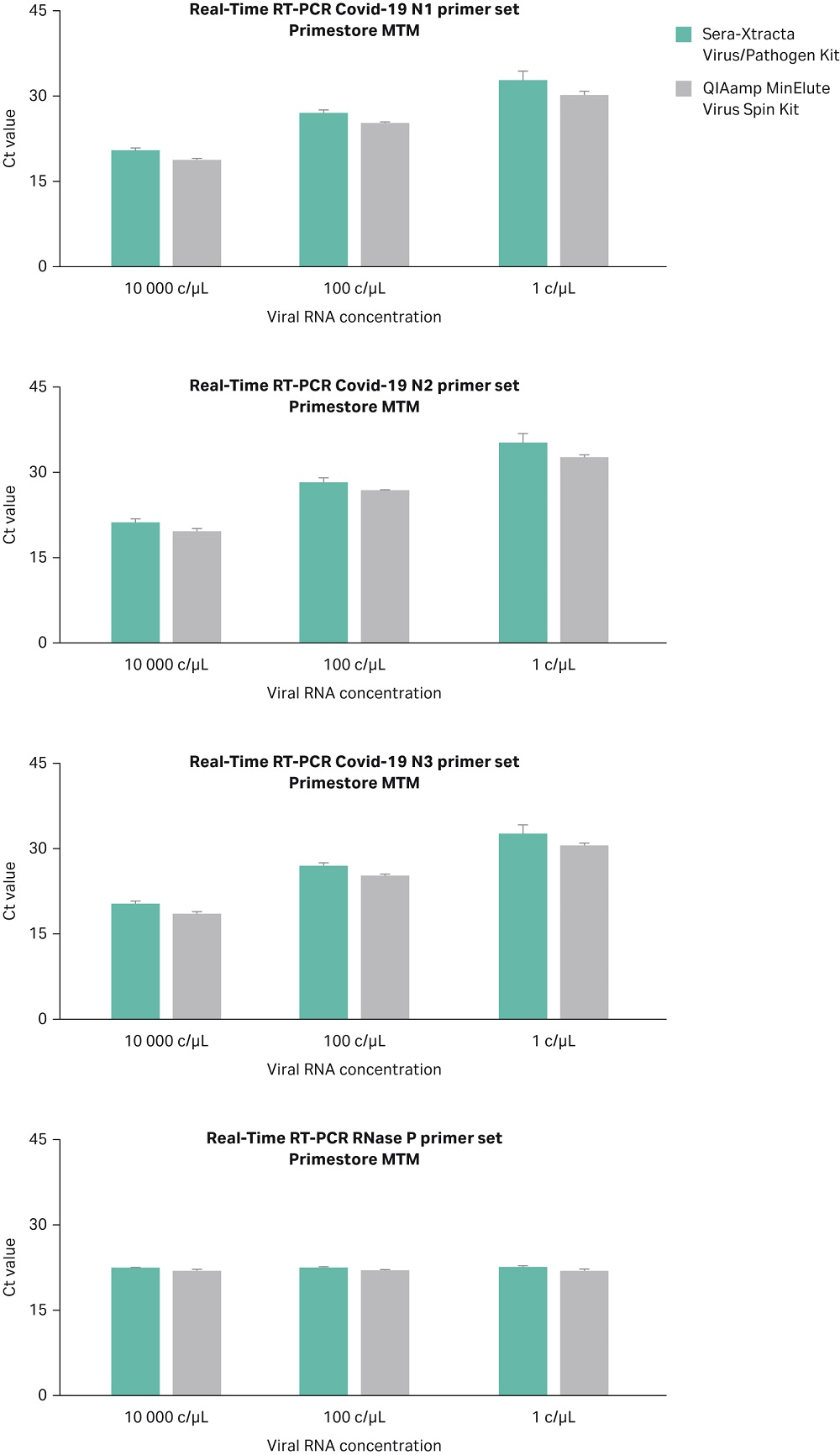
Fig 6. Ct values obtained for Sera-Xtracta Virus/Pathogen Kit and QIAamp Viral RNA Mini Kit for varying amounts of the viral synthetic RNA in the input sample as described on the graph. For clarity Ct values obtained for each of the three COVID-19 specific primer sets (N1, N2, N3) and human-specific RNase P primer set have been presented in separates graphs. Values averaged from three independent experiments, error bars represent standard deviation.
This data is based on a minimum of three independent experiments/replicate trials with the equal number of replicates in each experiment. All samples tested were treated equally (with the number of replicates being the same for all products tested in the comparison) and according to manufacturers’ protocol/recommendations. Data was collected at Cytiva, Maynard Centre, Cardiff, UK (R&D Laboratory) during March and April 2020 and is held at this location.
Automated versus manual protocol
The Sera-Xtracta Virus/Pathogen Kit has been assessed in the KingFisher™ DUO (ThermoFisher Scientific) automation system and results compared with the manual method. Experiments were carried out using 1, 100 and 10^4 copies/µL of synthetic SARS-CoV-2 RNA in PrimeStore MTM transport media containing human cells as described previously. Extracted samples were subjected to Real-Time RT-PCR following CDC protocol as described above. Data from two independent experiments from both manual and automated extraction methods are shown in Figure 7 and confirm that both methods enable detection of the viral RNA down to one copy/µL in the input sample. The automation script description is available upon request.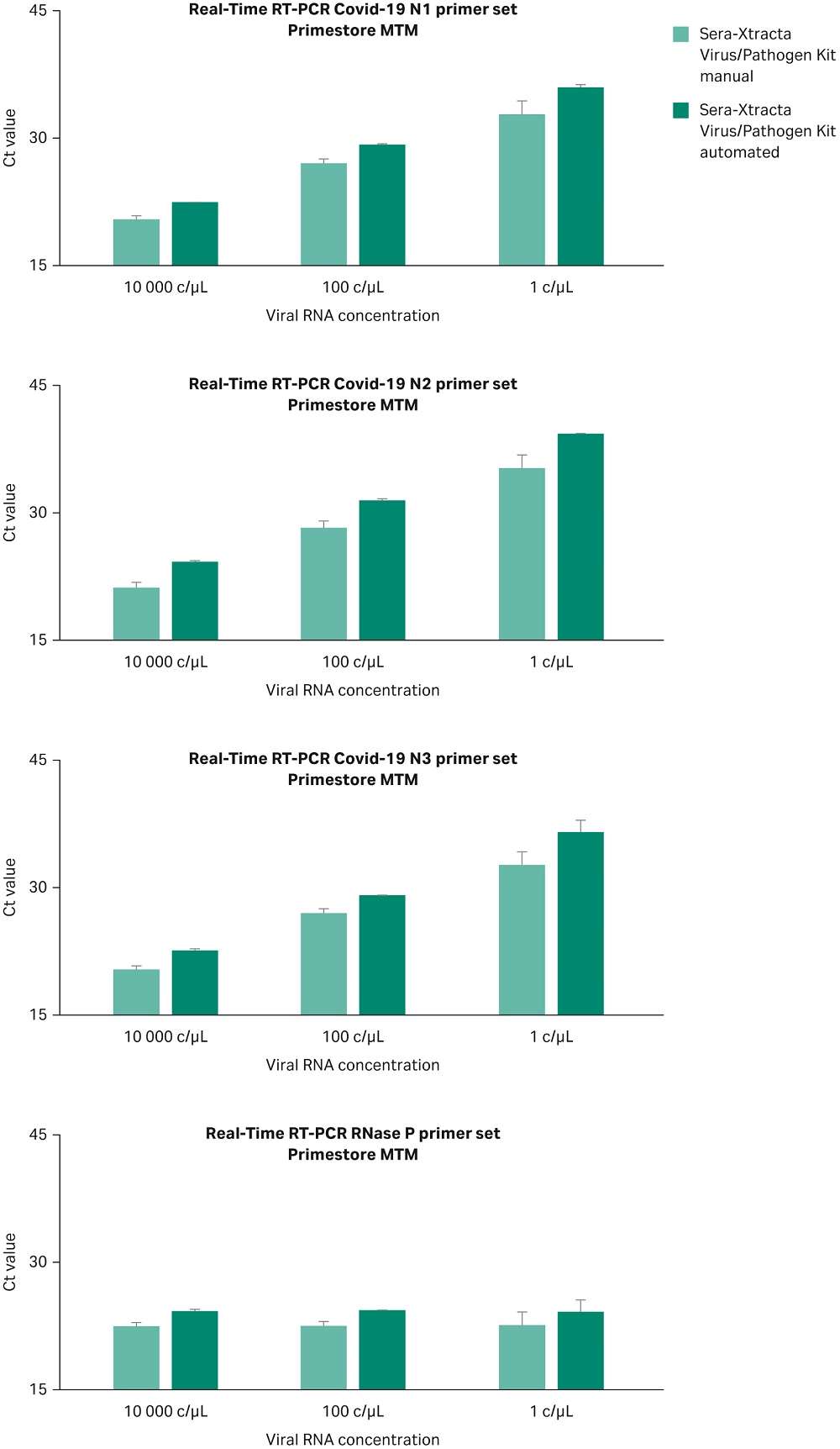
Fig 7. Ct values obtained for Sera-Xtracta Virus/Pathogen Kit in manual and automated protocol for varying amounts of the viral synthetic RNA with the input sample as described on the graph. For clarity Ct values obtained for each of the three COVID-19 specific primer sets (N1, N2, N3) and human-specific RNase P primer set have been presented in separates graphs. Values averaged from two independent experiments, error bars represent standard deviation.
The Sera-Xtracta Virus/Pathogen Kit has been optimized for viral nucleic acid extraction from PrimeStore MTM transport media. The performance of the kit was assessed using an alternative universal transport media (UTM) in both manual and automated protocol on KingFisher DUO. 200 µL of Copan UTM (COPAN Diagnostics Inc) containing human cells (~ 3x 10^5/mL) was spiked with a synthetic SARS-CoV-2 RNA at 1 and 10,000 copies/µL and processed as described before. Extracted samples were subjected to Real-Time RT-PCR following CDC protocol as described previously. Data from two independent experiments presented in Figure 8 confirms successful detection of the viral RNA load down to 100 copies/µL in the input sample independently of the method used (automation versus manual) when using alternative transport media.
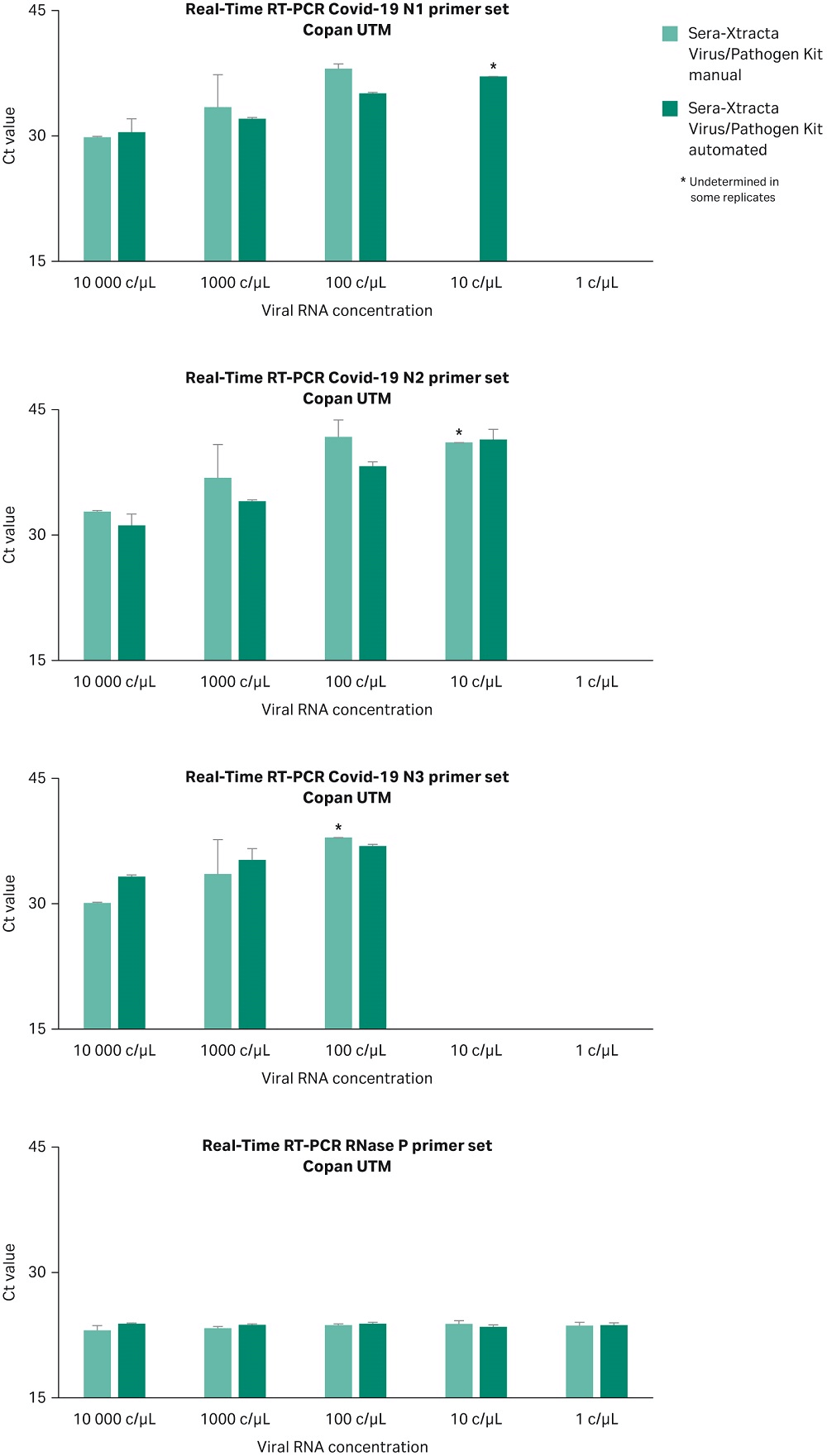
Fig 8. Ct values obtained for Sera-Xtracta Virus/Pathogen Kit in manual and automated protocol for varying amounts of the viral synthetic RNA in the COPAN UTM as described on the graph. For clarity Ct values obtained for each of the three COVID-19 specific primer sets (N1, N2, N3) and human-specific RNase P primer set have been presented in separates graphs. Values averaged from two independent experiments, error bars represent standard deviation. Please note that N3 primer set has been recently removed from CDC diagnostic panel but still included in RUO.
Conclusion
Sera-Xtracta Virus/Pathogen Kit gives efficient total nucleic acid (DNA/RNA) recovery from different sample types and in different concentrations with high sensitivity in downstream detection. The kit provides a simple and rapid method for molecular diagnostic laboratories looking to optimize their workflow for sensitive detection system for viruses and other pathogens found in low concentrations. Our data suggests that this kit and also the RNAspin Mini Kit can help address the demand for a universal extraction kit which copurifies both DNA and RNA or just RNA and can help address the current challenges faced with supplying kits for COVID-19 testing.
*Kit is for Research Use Only (RUO).
*Data for “Detection of Influenza A and SARS-CoV-2 from different sample types” provided by Luke T. Daum, Ph.D.
- Daum L, et al., (2007). Real-time RT-PCR assays for type and subtype detection of influenza A and B viruses. Influenza and Other Respiratory Viruses 1(4): 167-175.
- Centers for Disease Control and Prevention Division of Viral Diseases. Instructions for Use Catalog # 2019-nCoVEUA-01 1000 reactions, CDC-006-00019, Revision: 03.
https://www.fda.gov/media/134922/download. Published March 30, 2020. Accessed April 3, 2020. - Carrasco-Hernandez R, et al. (2017) Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review ILAR Journal 58(3): 343–358.