Introduction
Personalized medicine relies on accurate and sensitive profiling of tumor genetic make-up for targeted therapy decisions, monitoring the response to treatment and detecting changes in tumor molecular landscape that result in secondary resistance. Successful implementation of targeted therapy is dependent on both the availability of tumor tissue and on comprehensive biomarker profiling that reflects tumor heterogeneity. Liquid biopsies that rely on the presence of the genetic material released by cells into biofluids such as blood and urine are now entering routine clinical practice (1). As DNA present in the bloodstream represents a pool of genetic material released by all cells, these approaches are non-invasive, allow for molecular profiling in circumstances when tumor tissue biopsy is not feasible , and most importantly have the potential to fully uncover tumor heterogeneity and detect genetic alterations associated with tumor development.
Characteristics of cell-free DNA
The predominant size of cell-free DNA (cfDNA) circulating in the blood corresponds to the size of DNA wrapped around a histone octamer, i.e. ~ 170 bp. It is believed that the size of cfDNA is highly dependent on its source, with tumor-derived cfDNA exhibiting significant fragmentation and lower size profile when compared to healthy individuals (2,3). Based on those characteristics, any system that allows for enrichment of smaller, more degraded DNA fragments should be beneficial in detecting low frequency mutations associated with tumor evolution and therapy induced resistance. Similarly, any method of cfDNA extraction that effectively eliminates the contamination of high molecular weight (HMW) genomic DNA (gDNA) originating from white blood cells would be advantageous, removing the background signal that further dilutes tumor-derived cfDNA.
Sera-Xtracta™ Cell-Free DNA extraction kit
Sera-Xtracta™ Cell-Free DNA Kit* has been designed to effectively capture highly fragmented cfDNA and at the same time to minimize co-purification of higher molecular weight DNA. This unique formulation increases the probability of detecting mutations that constitute a minor proportion of genetic alterations present in the tumor. Identification of these mutations is critical for undertaking a fully informed decision regarding therapy profile. It also facilitates the detection of early resistance variants that call for a prompt change in the therapeutic approach to increase the chances of a successful outcome.
Cancer biomarker detection and mutation allele frequency in targeted next-generation sequencing (NGS)
Non-small cell lung cancer (NSCLC) is a leading cause of cancer mortality worldwide, mostly due to the fact that the diagnosis is being made at the very late stage with metastasis present. The five-year survival rate is lower than 15% (4). Current guidelines advocate the use of molecular profiling in the evaluation of genetic drivers in NSCLC and support the use of cfDNA-based profiling from patients with insufficient tissue (5). Endothelial growth factor receptor (EGFR) is the most well-established mutation with both prognostic and predicted value and with the approved Food and Drug Administration (FDA) therapy (6). Other well recognized genomic alterations associated with NSCLC, for which a targeted therapy approach is available, include rearrangements in anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 (ROS1), and ret proto-oncogene (RET) genes, B-Raf proto-oncogene (BRAF) mutation and met proto-oncogene (MET) amplification (7).
We have evaluated the performance of Sera-Xtracta™ Cell-Free DNA Kit using plasma from stage IV NSCLC patient samples.
Cell-free DNA yield and size profile
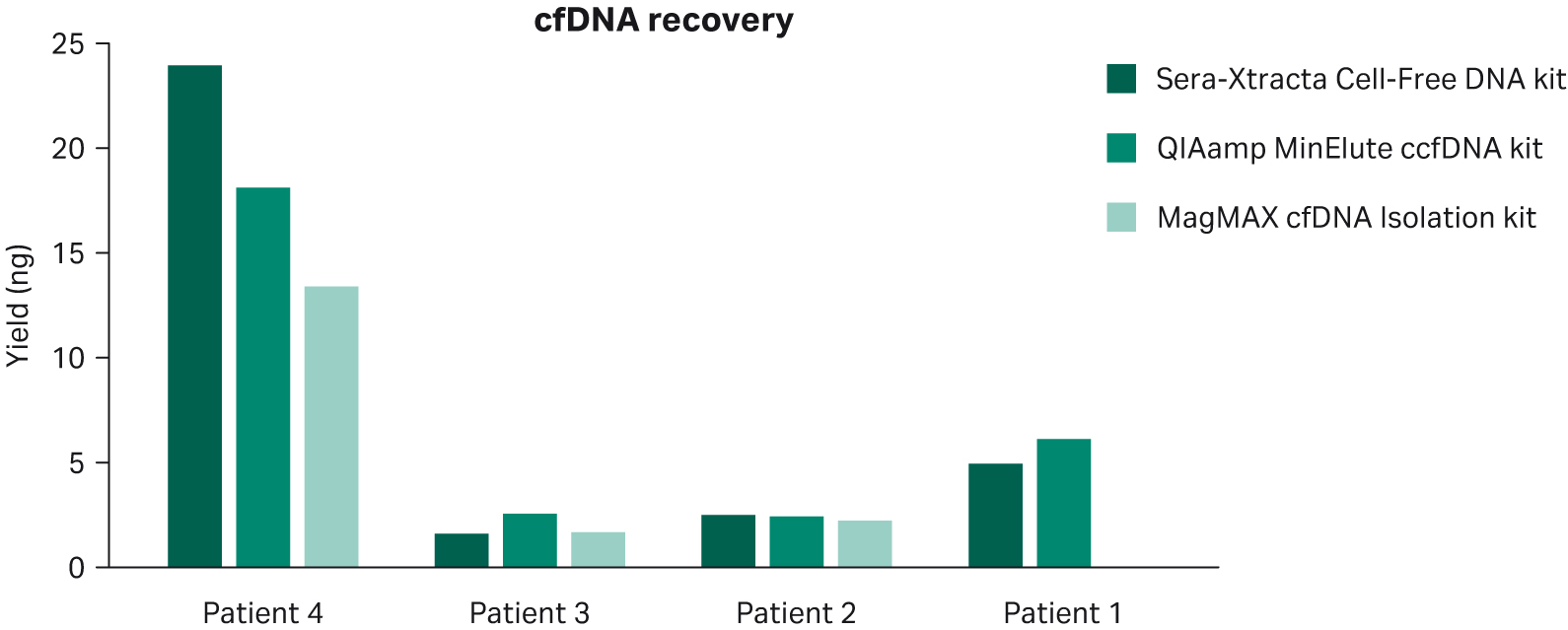
1 mL of plasma per patient obtained from blood collected in standard Ethylenediaminetetraacetic acid (EDTA) and Heparin blood collection tubes was processed using Sera-Xtracta™ Cell-Free DNA Kit, MagMAX™ cfDNA Isolation kit (all but patient 1) (ThermoFisher Scientific) and QIAamp™ MinElute™ ccfDNA kit (QIAGEN) following the manufacturers’ protocol. The results presented in Figure 1 show overall comparable performance for cfDNA recovery for all kits tested. No statistically significant difference was detected using mixed-effects model with Dunnett’s multiple comparison test.
Fig 1. cfDNA recovery from plasma following extraction with three different kits as described in the figure legend. Relative cfDNA yield and gDNA carry-over was calculated using smear analysis tool (2100 Expert software, Agilent) for fragments between 100-270 bp.
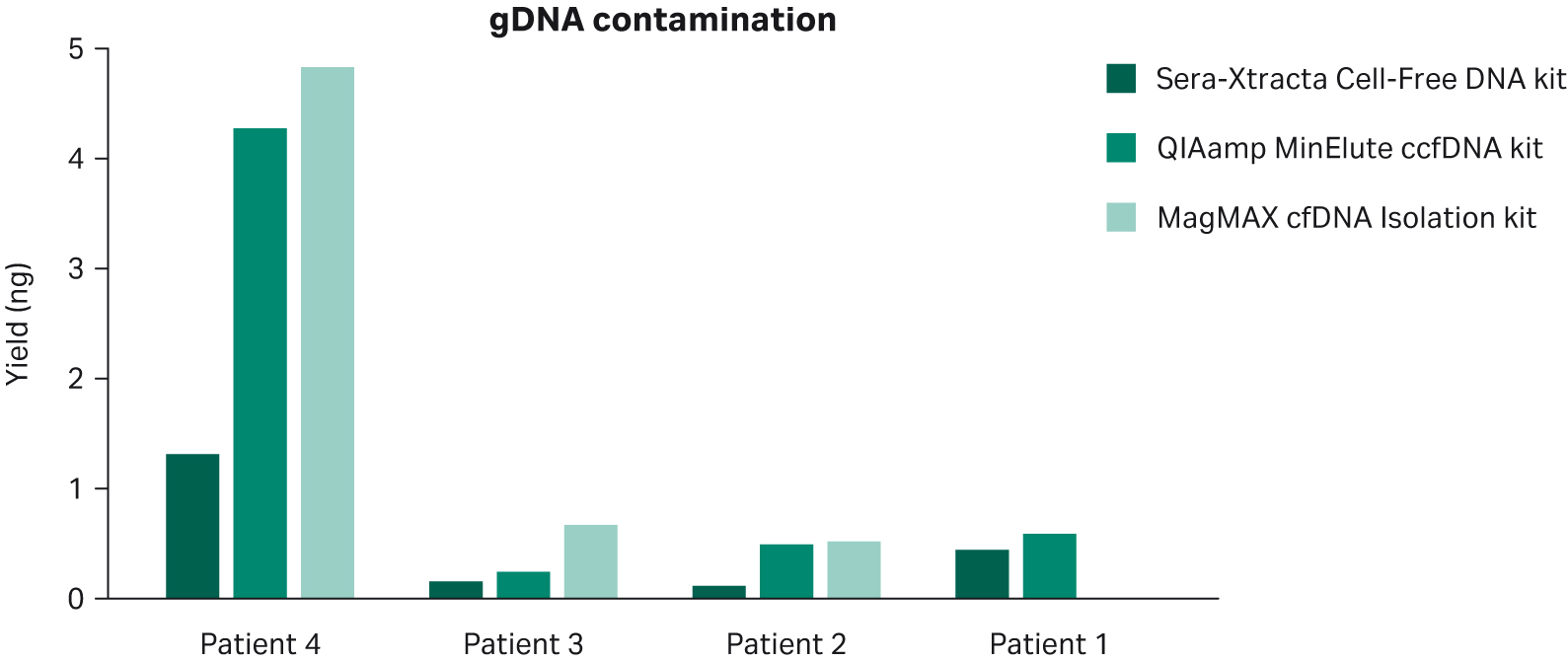
Data presented in Figure 2 indicates that Sera-Xtracta™ Cell-Free DNA Kit reduces the amount of gDNA carry-over in circumstances where a significant amount of white blood cell genetic material is released prior to plasma processing, as apparent for Patient 4. Note that no statistically significant difference was detected using mixed-effects model with Dunnett’s multiple comparison test.
Fig 2. Carryover of HMW DNA from plasma following extraction with three different kits as described in the figure legend. Relative HMW DNA yield was calculated using smear analysis tool (2100 Expert software, Agilent) for fragments above 700 bp.
Cancer biomarker detection and mutation allele frequency in targeted NGS
Extracted cfDNA was concentrated to 10.4 μL to allow for maximum input into library preparation. Samples were tested using the Target Selector™ NGS Lung Panel service (Biocept) and run alongside positive controls (Table 1). The panel allows for preparation of amplicon-based NGS libraries to detect somatic alterations in 12 clinically-relevant lung cancer genes. Panel list and limit of detection (LOD) can be found in Table 2 and Table 3 respectively.
Table 1 Target Selector NGS Lung Panel positive controls run alongside clinical samples
| Sample ID | SNVs and INDELs | CNVs | Fusions | |||||
|---|---|---|---|---|---|---|---|---|
| Gene | MAF % | AA Chg | Gene | Gain / Loss | CNV ratio | Variant (exons) | Mol Cov. Mutant | |
| Positive control | NRAS | 0.5249 | p.Q61R | MET | Gain | 1.34 | not determined | |
| ALK | 0.4327 | p.G1202R |
||||||
| ALK | 0.5446 | p.F1174L |
||||||
| PIK3CA | 0.8436 | p.H1047R |
||||||
| EGFR EGFR |
0.887 0.2545 |
p.L747_P753delinsS p.T790M |
||||||
| EGFR | 0.536 | p.L858R |
||||||
| BRAF | 0.4318 | p.V600E |
||||||
| KRAS | 0.3642 | p.Q61H |
||||||
| KRAS | 0.7926 | p.G12D |
||||||
| ERBB2 | 1.3353 | p.A771_Y772insYVMA |
||||||
Table 2 Target Selector NGS Lung Panel gene list. Note that genes indicated in bold are referenced in National Comprehensive Cancer Network (NCCN) Guidelines and are targeted by FDA-approved therapy.
| Target selector NGS Lung Panel gene list | |||||
|---|---|---|---|---|---|
| Hotspot genes | CNVs | Fusions | Exon variants | ||
| ALK | KRAS | PIK3CA | ALK | ||
| BRAF | MAP2K1 | ROS1 | MET | RET | MET exon 14 skipping |
| EGFR | MET | TP53 | ROS1 | ||
| ERBB2 | NRAS | ||||
Table 3 Target Selector ™ NGS Lung Panel LOD
| Target selector NGS Lung Panel content | |
|---|---|
| Assay input | DNA + RNA |
| Hotspot SNV/indel LOD | 0.1% MAF |
| De novo LOD | 0.5% MAF |
| CNV LOD | 1.12X |
| Fusion/exon skipping LOD | 3 molecular counts |
The results presented in Table 4 show that samples extracted with Sera-Xtracta™ Cell-Free DNA Kit exhibit higher mutation allele frequency (MAF) in all samples tested, which is indicative of enrichment in tumor-derived fraction.
Table 4 The summary of MAF detected using Target Selector NGS Lung Panel for NCSC samples. Note that Patient 3 plasma yielded a suboptimal level of cfDNA for the NGS library prep that might account for the discrepancy in allele variant and MAF (i.e. not detected for MagMAX™ cfDNA Isolation kit).
| Patient 4 BRAF V600E |
Patient 3 TP53 |
Patient 2 BRAF V600E |
Patient 1 TP53 G245A |
|
|---|---|---|---|---|
| Sera-Xtracta™ Cell-Free DNA Kit |
0.64% |
0.61% (Variant ID: R282W) |
2.15% |
2.14% |
| QIAamp™ MinElute™ ccfDNA Kit |
0.27% |
0.35% (Variant ID: C277Y) |
0.26% |
1.46% |
| MagMAX™ cfDNA Isolation Kit |
0.27% |
Not detected |
0.32% |
N/A |
Patients 2 and 4 were both identified as positive for mutation in BRAF gene at position 600, in which substitution of valine by glutamic acid leads to constitutively active Raf kinase and uncontrolled growth (6). The mutation is consistently detected in all samples processed with all kits; however, MAF, which is indicative of the relative content of tumor-derived cfDNA fraction, is consistently higher in samples processed with Sera-Xtracta™ Cell-Free DNA Kit. The advantage of minimizing gDNA carry-over is particularly evident for Patient 2, where the relative recovery of cfDNA (Fig 1) is similar between all kits tested but the amount of HMW contamination is considerably lower for samples extracted with the Sera-Xtracta™ kit. This results in at least 6.7 x higher frequency of mutated allele detected.
Patients 1 and 3 were identified positive for mutation in tumor protein 53 (TP53), which is one of the most frequently mutated genes in human cancers and encodes tumor suppressor protein p53. Alterations in TP53 gene are found in 35% to 60% of NSCLC patients, more frequently in squamous cell carcinomas and patients with a smoking history (8). P53 is composed of three distinct domains with the DNA binding domain being key in tumor-suppressing function of the protein and the one representing mutation hotspot. 90% of point mutations occur in highly conservative sites including 175, 245, 248, 249, 273, 282 (8). Two mutations in this hotspot region were identified in cfDNA extracted with the Sera-Xtracta™ kit (G245A and R282W in Patient 1 and Patient 3 respectively). Surprisingly, the results for Patient 3 were inconsistent when comparing samples processed with competitor kits: sample extracted with MagMax™ cfDNA Isolation kit failed to yield a positive signal for any of the cancer-associated mutations tested, while sample processed with QIAamp™ MinElute™ ccfDNA kit detected an alternative TP52 mutation, i.e. C277Y. This mutation is not recognized as a hotspot single-nucleotide variant (SNV) and lies below the LOD of Target Selector NGS Lung Panel specified for de novo SNV (> 0.5%). As such, it would be interpreted as an inconclusive result.
Concluding remarks
The data presented strongly suggests that size selection-based extraction of cfDNA offers a distinct advantage in clinically relevant scenarios. Enrichment of tumor-derived fraction allows for detection of low frequency mutations that otherwise might be missed with standard extraction methods. The clear advantage of the Sera-Xtracta™ technology can be attributed to the three key features: efficient extraction of the main cfDNA peak, reduction in gDNA carry-over, and increased recovery of small fragments.
This data is based on three independent experiments with the equal number of replicates in each experiment and a fourth experiment with two replicates. All samples tested were treated equally (with the number of replicates being the same for all products tested in the comparison) and according to manufacturers’ protocol and recommendations. cfDNA extraction was carried out at Cytiva, The Maynard Centre, Forrest Farm Ind. Estate, Longwood Drive, Cardiff CF14 7YT (R&D Laboratory) during August 2019. NGS library prep, NGS and data analysis were performed at BioCept Inc. 5810 Nancy Ridge Dr # 150, San Diego, CA 92121, United States (R&D Laboratory) during December 2019 and data is held at this location.
Learn more about working with magnetic beads
*For research use only (RUO). Not for diagnostic use.
- David S. Ettinger, MD, of The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins
- Mouliere F, R. B. (n.d.). Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumor-derived circulating DNA. PLoS One. 2011;6(9):e23418. doi:10.1371/journal.pone.0023418.
- Sanchez, C. S. (n.d.). New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. npj Genomic Med 3, 31 (2018). https://doi.org/10.1038/s41525-018-0069-0.
- Majem M, R. J. (n.d.). Tumor heterogeneity: evolution through space and time in EGFR mutant non small cell lung cancer patients. . Transl Lung Cancer Res. 2013;2(3):226-237. doi:10.3978/j.issn.2218-6751.2013.03.09.
- Pritchett Michael A., D. R.-N. (n.d.). Prospective Clinical Validation of the InVisionFirst-Lung Circulating Tumor DNA Assay for Molecular Profiling of Patients With Advanced Nonsquamous Non–Small-Cell Lung Cancer. JCO Precision Oncology 2019 :3, 1-15.
- Back Robert C., H. K. (n.d.). NSCLC: An Update of Driver Mutations, Their Role in Pathogenesis and Clinical Significance. Rhode Island Medical Journal.
- American Cancer Society. (2019). Treating Non-Small Cell Lung Cancer. cancer.org | 1.800.227.2345.
- Hou H, Q. K. (n.d.). Concurrent TP53 mutations predict poor outcomes of EGFR-TKI treatments in Chinese patients with advanced NSCLC. . Cancer Manag Res. 2019;11:5665-5675. Published 2019 Jun 21. doi:10.2147/CMAR.S201513.