In 2019, a novel coronavirus (SARS‑CoV-2) was identified as the source of a pneumonia outbreak in Wuhan, China. The extraordinarily rapid spread of this disease, termed COVID-19, triggered numerous and urgent challenges to health systems, diagnostic laboratories and basic research communities throughout the world. Demand for reproducible and fast pathogen nucleic acid extraction has been unprecedented and as a result has become a limiting factor for diagnostic laboratories and research institutions globally.
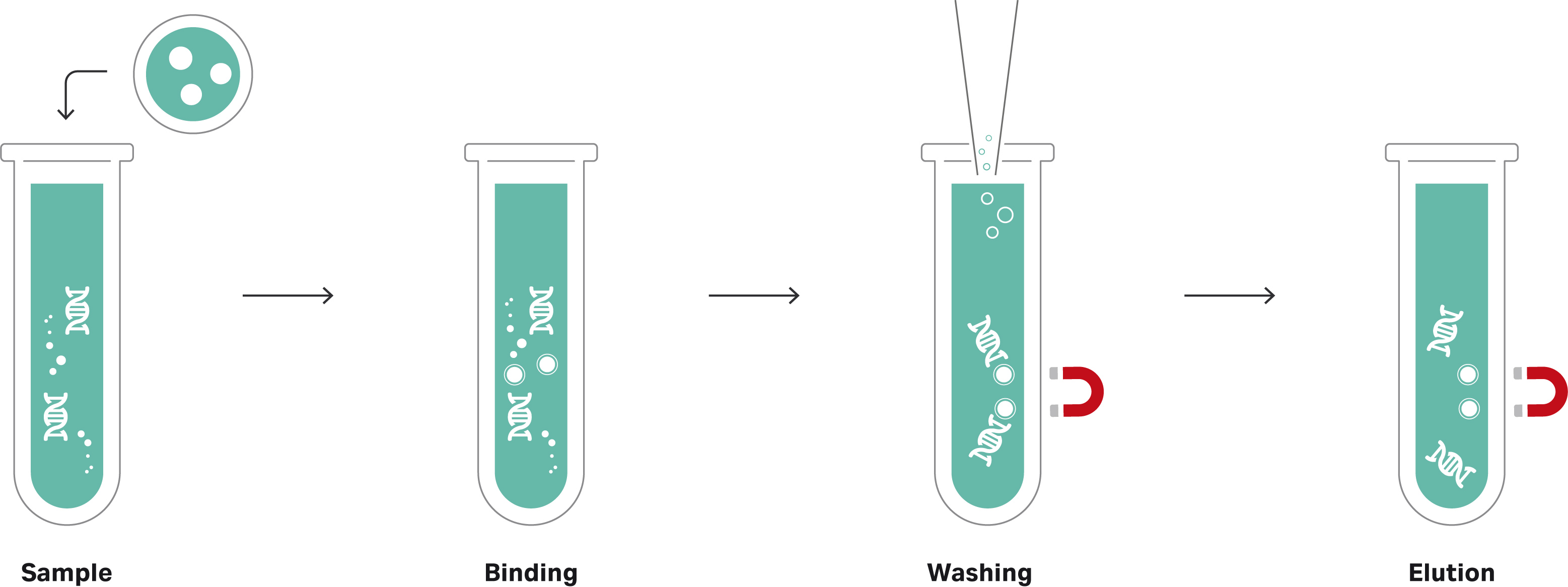
The Sera-Xtracta Virus/Pathogen Kit provides a simple and rapid method to optimize the workflow for sensitive detection of viruses and other pathogens found in low concentrations. The silica coated magnetic bead-based automatable kit can be used with a range of sample types including respiratory biological matrices, blood and universal transport media. Using non-carrier RNA based chemistry, the kit is designed for high-throughput selective binding of total nucleic acid from bacteria and viruses, including: Adenovirus (Type 14), Influenza A (H3N2) and SARS-CoV-2 (COVID-19). The isolation procedure illustrated in Figures 1a and 1b can be completed in less than 60 minutes (‘hands-on’ time) for reproducible yields compatible with molecular biology techniques, including quantitative polymerase chain reaction (qPCR) Real-Time quantitative polymerase chain reaction (RT-qPCR), droplet digital PCR (ddPCR), and next-generation sequencing (NGS) applications.
Filter papers, such as Whatman 903 Proteinsaver cards, have been used for the collection, storage, and testing of blood specimens in the diagnostic screening of metabolic and inherited disorders in newborn babies in the United States for many years. More recently, many countries have been using them for HIV related molecular assays, including early infant diagnosis using Roche Amplicor™ HIV-1 DNA PCR testing (supported by the U.S. President’s Emergency Plan for AIDS Relief [PEPFAR]), real-time PCR-based HIV-1 diagnosis and viral load measurement, and HIV-1 drug resistance genotyping (1), due to attributes such as its low cost, the minimally invasive procedure, the simplicity of transportation, collection and storage and the reduced biohazard risks (2).
Samples from suspected cases of COVID-19 are typically taken by swab and stored in universal transport media (UTM) for stabilization and transportation prior to testing. In some cases, saliva samples are collected (3). One key advantage of collecting and storing UTM or biofluids on Whatman 903 cards is that once completely dried, samples are considered non-infectious and non-hazardous(4, 5) allowing them to be shipped safely with no cold chain storage required for preservation, minimizing the need for specialized transportation requirements (Table 1). In addition, a recent study found that, on cardboard, no viable SARS-CoV-2 was measured after 24 hours (6). This is a good gauge for other porous material like paper, indicating 903 cards could provide a safe transportation method for COVID-19 samples.
The following data demonstrates proof of concept for using the Sera-Xtracta Virus/Pathogen Kit for the extraction of viral RNA from UTM and biofluids collected on Whatman 903 cards.
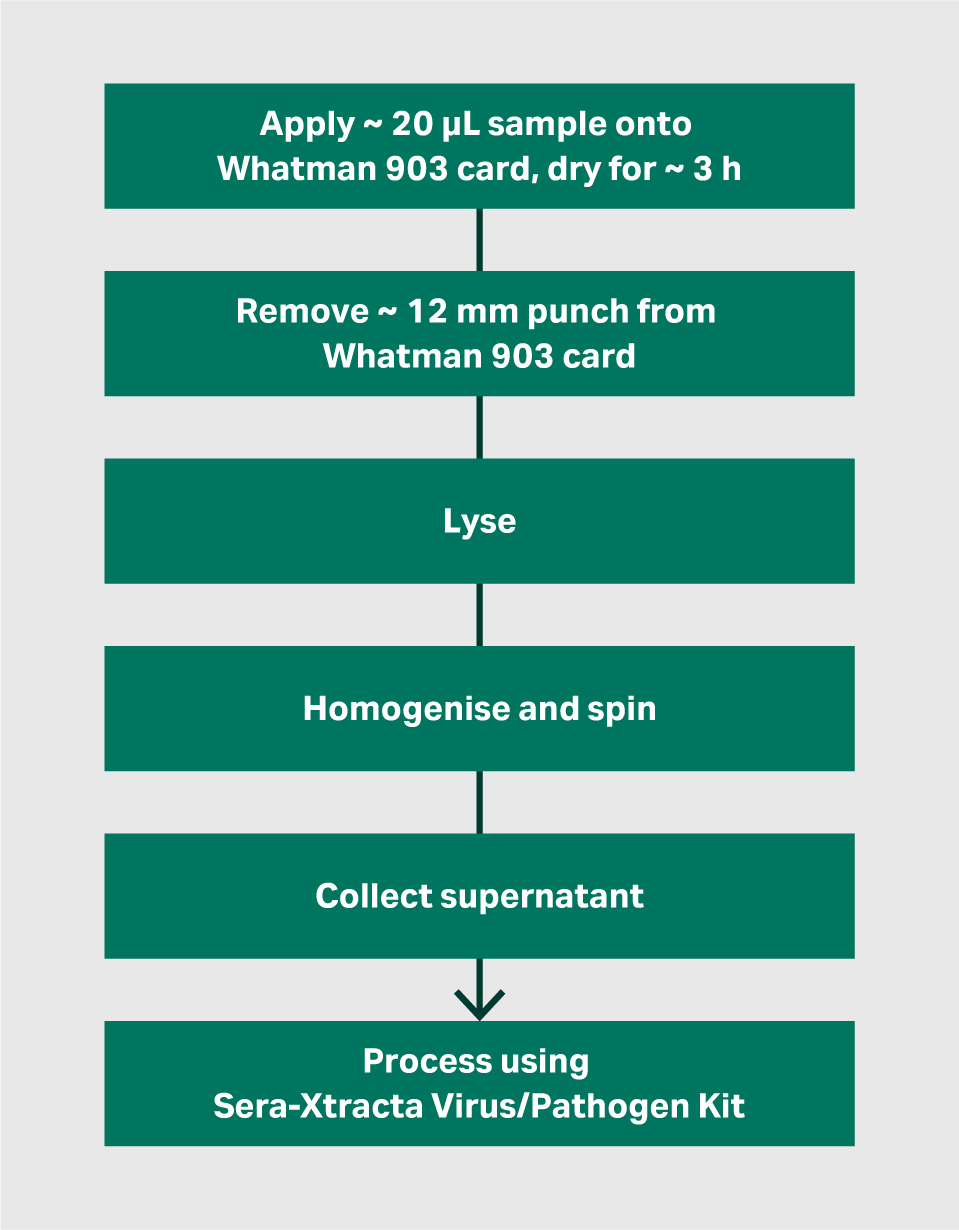
Figure 1a Workflow for initial lysis and homogenisation of Whatman 903 cards
Figure 1b Workflow using Sera-Xtracta Virus/Pathogen kit
Experimental set up
The Sera-Xtracta Virus/Pathogen Kit (product code 29506009) was used to extract SARS-CoV-2 (COVID-19) RNA from UTM and blood collected on Whatman 903 cards in an experimental setting, highly relevant to virus detection from human swab samples or biofluids (such as blood or saliva) routinely performed in a diagnostic laboratory.
The experiments used heat inactivated SARS-CoV-2 viral particles (American Type Culture Collection, ATCC) spiked at 50 copies/µL into universal transport media (COPAN Diagnostics Inc.) containing human cells (~ 3 × 10^5/mL) to mimic a clinical swab sample. In addition, viral particles were spiked into whole human blood to examine storage and extraction of COVID-19 RNA from a human biofluid. 20 µL aliquots of spiked blood and spiked UTM/cell samples were spotted onto Whatman 903 cards and left to dry at room temperature for 3 hours, then stored in a Whatman foil barrier zip sealing bag (product code 10534321). 20 µL aliquots of un-spiked cell suspension and whole blood (no viral RNA) were spotted onto Whatman 903 cards and served as assay controls to eliminate the possibility of false positives.
Viral RNA was extracted from spotted Whatman 903 cards following overnight (T0) and 14 days (T14) storage at room temperature. Punches of ~ 12 mm diameter (i.e., sufficient to remove the entire 20 µL dried sample) were removed from each card and subjected to an initial lysis step (refer to ‘Methods’ section) prior to processing using the Sera-Xtracta Virus/Pathogen kit according to the Manufacturer’s Instructions.
20 µL aliquots of UTM containing human cells and 20 µL aliquots of whole blood spiked with viral RNA at 50 copies/µL were added to 300 µL aliquots of nuclease free water and extracted using the standard Sera-Xtracta Virus/Pathogen Kit protocol. These served as liquid controls.
Samples were eluted in 50 µL of nuclease-free water and subjected to Real-Time RT-PCR assay following the Centres for Disease Control and Prevention (CDC) guidelines(7). Briefly, extracted samples (5 µL of eluant per well) were run in technical duplicates using TaqPath™1-Step RT-qPCR Master Mix, CG (ThermoFisher Scientific) and CDC 2019 Novel Coronavirus (2019-nCoV) Diagnostic Panel primers (N1, targeting a region of COVID-19 nucleocapsid gene and RNase P, targeting human RNase P gene, IDT).
As per CDC guidelines, only samples for which the N1 marker crossed the threshold at Ct < 40 were considered positive; no amplification in the N1, COVID-19 specific primer set were observed in un-spiked controls (undetermined, data not shown).
Methods
Reagents were prepared as described in the Sera-Xtracta Virus/Pathogen kit user manual.
Lysis and homogenisation of Whatman 903 cards - The following steps should be performed prior to using the Sera-Xtracta Virus/Pathogen Kit when extracting viral RNA from Whatman 903 cards.
- Remove a punch of ~ 12 mm diameter (i.e., sufficient to remove 20 µL dried sample) from the Whatman 903 card and place into a sterile, nuclease free 1.5 mL Eppendorf™ tube.
- Add 10 µL Proteinase K (@ 20 mg/mL, supplied in Sera-Xtracta Virus/Pathogen Kit) and 500 µL lysis buffer (7 M Guanidine hydrochoride, 50 mM Tris-HCl, 10 mM EDTA, 5% w/v TWEEN-20, 4% w/v Triton X-100, adjusted to pH 7.0) to each tube.
- Incubate on a thermomixer at 37°C for 1 hour, shaking at 450 rpm.
- Post incubation, using a sterile 1 mL pipette tip, mash / break up the punch to obtain a pulp-like slurry.
- Centrifuge at ~ 12 000 × g for 4 minutes.
- Using a pipette tip, move the pulp from the punch to the side of the tube (above the supernatant) and squeeze out the liquid by pressing the punch/pulp against the side of the tube with the 1 mL pipette tip.
- Leave the tube to stand for ~ 5 minutes to allow supernatant to drip out of the punch.
- Remove supernatant to a fresh sterile Eppendorf tube and process up to 400 µL of the 903 extract as described in the Sera-Xtracta Virus/Pathogen Kit user manual (product code 29506009).
Results and Discussion
Detection of SARS-CoV-2 RNA at 50 copies/µL in the input sample
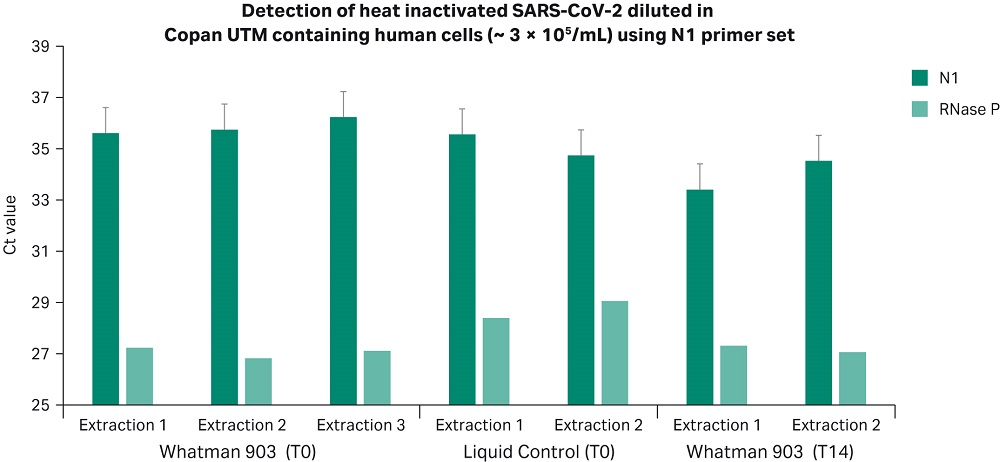
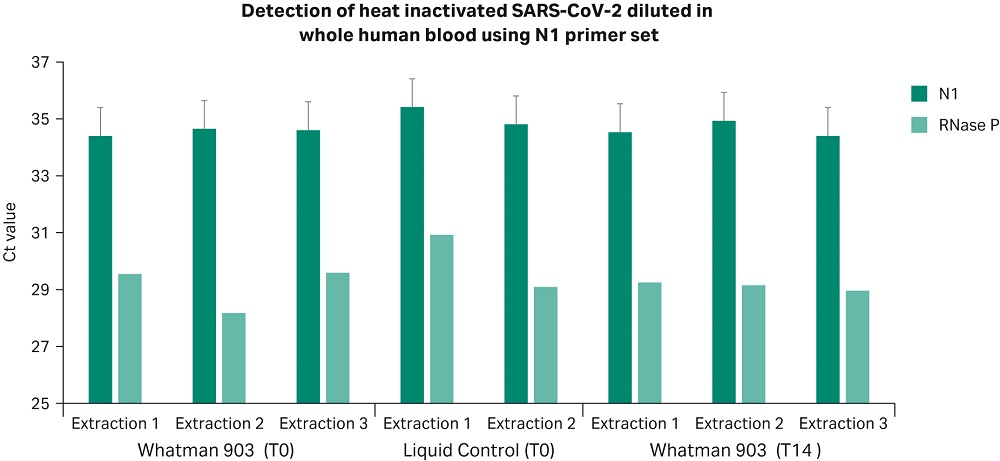
Results presented in Figures 2 and 3 confirm that Whatman 903 cards and the Sera-Xtracta Virus/Pathogen Kit provide reliable sample storage and nucleic acid purification (NAP) chemistry for the detection of viral RNA down to 50 copies/µL in whole blood and UTM containing human cells following room temperature storage for 14 days. Ct values at T0 and T14 were comparable to liquid controls (p > 0.05, unpaired t-test. Note: undetermined Ct values not included in t-test), demonstrating storage of COVID-19 viral RNA on Whatman 903 cards resulted in no loss in yield.
Whatman 903 cards are a viable option for the collection, storage and transportation of viral RNA and COVID-19 samples. Stability of COVID-19 RNA on Whatman 903 cards for 14 days provides sufficient time for transportation to the laboratory for testing. For the successful storage of HIV viral RNA, the WHO recommend dried blood spots (i.e., the collection of blood on a paper card, such as Whatman 903, and subsequently using the dried blood spots (DBS) for diagnostic purposes) can be kept and/or transported at ambient temperature up to 14 days after collection. After this time, DBS must be either processed for genotyping or frozen at -20°C or below (8). A list of publications studying longer storage conditions for viral RNA (HIV) stored on Whatman 903 cards and their main findings is presented in Table 1.
Figure 2
Figure 3
Figures 2 and 3. Ct values obtained for viral RNA spiked into UTM containing human cells (Figure 2) and whole human blood (Figure 3) and stored at room temperature on Whatman 903 cards for 24 hours (T0) and 14 days (T14). Ct values obtained for the N1 COVID-19 specific primer set with 50 copies/µL (1000 copies/extraction) of the heat inactivated viral RNA in the input sample and human-specific RNase P primer set are presented. Values for the N1 COVID-19 specific primer set are averaged from two technical replicates at T0 and three technical replicates at T14 (for each independent extraction). Error bars represent standard deviation. Note that COVID-19 specific amplification was not detected in any of the un-spiked control samples (Ct undetermined), data not shown.
Table 1. Overview of published studies investigating optimal storage conditions for Whatman 903 cards spotted with dried blood spots (DBS) for HIV viral RNA genotyping (taken from the WHO Manual for HIV Drug Resistance Testing Using Dried Blood Spot Specimens (8)).
| Storage conditions tested | ||||
|---|---|---|---|---|
| Study | Time | Temperature/Humidity | Desiccant | Outcomes |
| Garcia-Lerma 9 | 1–16 weeks | 37°C / high humidity, -20°C | Yes | DBS stable at 37°C only for 1-2 weeks. -20°C recommended for long-term storage. -20°C superior for short and long term. |
| Buckton 10 | 3 months | -20°C, 4°C, 20°C | Yes | HIV DNA PCR only. No observed degradation in HIV DNA during 3 month study period. |
| Bertagnolio 11 | 3 months | 37°C / 85% humidity | Yes | Good amplification rate (90%) |
| McNulty 12 | 6 years 5 years 2–3 years |
-30°C Ambient temperature and -70°C -20°C |
Yes | Complete degradation at ambient temperature, stable at -30°C and -70°C; -20°C recommended for long-term storage. |
| Nelson 13 | 3–6 years | Ambient temperature | Yes | Moderately successful amplification rate (69%); 1 log drop in viral load. |
| Wallis 14 | 3 months | Ambient temperature, 4°C, -20°C | Yes | Some reduction in amplification rate at ambient temp. vs. 4°C or -20°C. |
Conclusions
The Sera-Xtracta Virus/Pathogen kit provides efficient total nucleic acid (DNA/RNA) recovery from different sample types and in different concentrations with high sensitivity in downstream detection. The kit provides a simple and rapid method for molecular diagnostic laboratories looking to optimize their workflow for sensitive detection system for viruses and other pathogens found in low concentrations. It is compatible with automation and being based on magnetic beads allows for considerable instrumentation flexibility.
The kit has been developed in response to a global COVID-19 pandemic and aimed to address current challenges faced with supplying kits for testing and scientific research. The format of the kit and a rapid protocol provide an excellent high-throughput solution that should aid in global efforts of scientific and medical community to detect and study the virus.
The Sera Xtracta Virus/Pathogen kit also provides a reproducible method for pathogen nucleic acid extraction from Whatman 903 cards. One key advantage of collecting and storing UTM or biofluids on Whatman 903 cards is that once completely dried, samples are considered non-infectious and non-hazardous(4, 5) allowing them to be shipped safely, with no cold chain storage required for preservation, minimizing the need for specialised transportation requirements. This study demonstrates Whatman 903 cards are a viable option for the collection, storage and transportation of viral RNA and COVID-19 samples, providing confident and reliable extraction of viral RNA and detection down to 50 copies/µL from whole blood and UTM containing human cells. Viral RNA stored on Whatman 903 cards was stable at room temperature for 14 days, therefore providing sufficient time for transportation to the laboratory for testing.
The Sera-Xtracta Virus/Pathogen kit is for research use only (RUO).
Learn more about Sera-Xtracta Virus/Pathogen Kit
- Yang, C. et al. Development and Application of a Broadly Sensitive Dried-Blood-Spot-Based Genotyping Assay for Global Surveillance of HIV-1 Drug Resistance. J Clin Microbiol. 2010. 48(9):3158-64. doi: 10.1128/JCM.00564-10.
- Dried Blood Spot Collection Cards Market Size, Market Research and Industry Forecast Report, 2026 (Includes Business Impact of COVID-19). https://www.trustedbusinessinsights.com/details/dried-blood-spot-collection-cards-market-size-market-research-and-industry-forecast-report-2026-tbimrg20a408 Accessed July 2020.
- U.S. Food & Drug Administration News Release. May 08, 2020. Coronavirus (COVID-19) Update: FDA Authorizes First Diagnostic Test Using At-Home Collection rel="noopener noreferrer" of Saliva Specimens https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-diagnostic-test-using-home-collection-saliva. Accessed July 2020.
- World Health Organization. 2013. Guidance on regulations for the Transport of Infectious rel="noopener noreferrer" Substances 2013-2014. World Health Organization. https://apps.who.int/iris/bitstream/handle/10665/78075/WHO_HSE_GCR_2012.12_eng.pdf;jsessionid=B683D5A76E94D16B1B7945A9437E8E03?sequence=1 Accessed June 2020.
- Parry, C. M. et al. Field Study of Dried Blood Spot Specimens for HIV-1 Drug Resistance Genotyping. J Clin Microbiol. 2014. 52(8):2868-75. doi: 10.1128/JCM.00544-14.
- Hammett, E. How long does Coronavirus survive on different surfaces?. BDJ Team 7, 14–15 (2020). rel="noopener noreferrer" https://doi.org/10.1038/s41407-020-0313-1 Accessed June 2020.
- https://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html Accessed June 2020.
- World Health Organization. rel="noopener noreferrer" 2010. WHO Manual For HIV Drug Resistance Testing Using Dried Blood Spot Specimens: https://www.who.int/hiv/topics/drugresistance/dbs_protocol.pdf Accessed June 2020.
- Garcia-Lerma, J. G. et al. Rapid decline in the efficiency of HIV drug resistance genotyping from dried blood spots (DBS) and dried plasma spots (DPS) stored at 37°C and high humidity. 2009. J Antimicrob Chemother.
- Buckton, A. J. et al. Development and optimization of an internally controlled dried blood spot assay for surveillance of human immunodeficiency virus type-1 drug resistance. 2008. J Antimicrob Chemother 62, 1191-1198.
- Bertagnolio, S. et al. HIV-1 drug resistance surveillance using dried whole blood spots. 2007. Antivir Ther 12, 107-113.
- McNulty, A. et al. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. 2007. J Clin Microbiol 45, 517-521.
- Nelson, J. A. et al. Nevirapine resistance in human immunodeficiency virus type 1-positive infants determined using dried blood spots stored for up to six years at room temperature. J Clin Microbiol 47, 1209-1211 (2009).
- Wallis, C. L., Bell, C. M., Horsfield, P., Rinke de Wit, T. & Stevens, W. Affordable resistance test for Africa (ARTA): DBS storage and extraction conditions for HIV subtype C. Abstract WEPEB214. The 5th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention (Cape Town, South Africa; July 19-22, 2009).
Detection and genotyping of Viral RNA
- Detection of hepatitis C virus RNA in dried blood spots, http://priede.bf.lu.lv/ftp/grozs/Mikrobiologijas/Virusol/2014%20R/Birkava_HCV%20asins%20traipos.pdf
- Simple and Reliable Method for Detection and Genotyping of Hepatitis C Virus RNA in Dried Blood Spots Stored at Room Temperature, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC130818/
- Dried blood spot self-sampling at home is a feasible technique for hepatitis C RNA detection, https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0231385