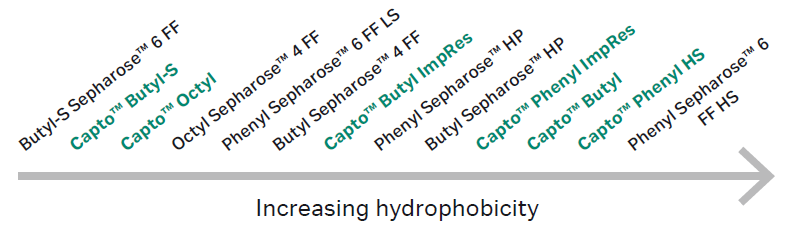
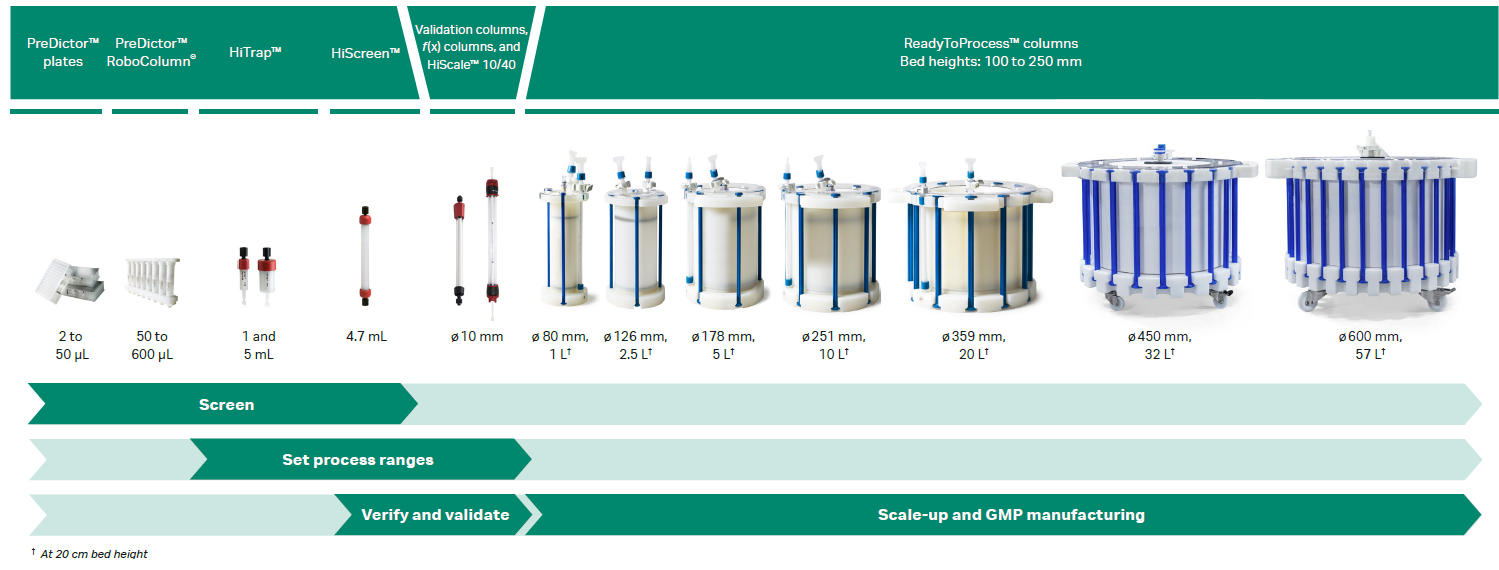
What is hydrophobic interaction chromatography?
Hydrophobic interaction chromatography (HIC) separates proteins according to differences in their surface hydrophobicity. HIC utilizes a reversible interaction between the proteins and the hydrophobic ligand of a HIC resin.
The interaction between hydrophobic proteins and a HIC resin is greatly influenced by the running buffer. A high salt concentration enhances the interaction. Lowering the salt concentration weakens the interaction.
How does hydrophobic interaction chromatography work?
Proteins with different degrees of surface hydrophobicity can be separated using hydrophobic interaction chromatography. The proteins are bound to the hydrophobic ligand on the HIC resin in a binding buffer with a high salt concentration.
When the ionic strength of the buffer is reduced, the interaction is reversed. Therefore, the protein with the lowest degree of hydrophobicity is eluted first. The most hydrophobic protein elutes last, requiring a greater reduction in salt concentration to reverse the interaction.
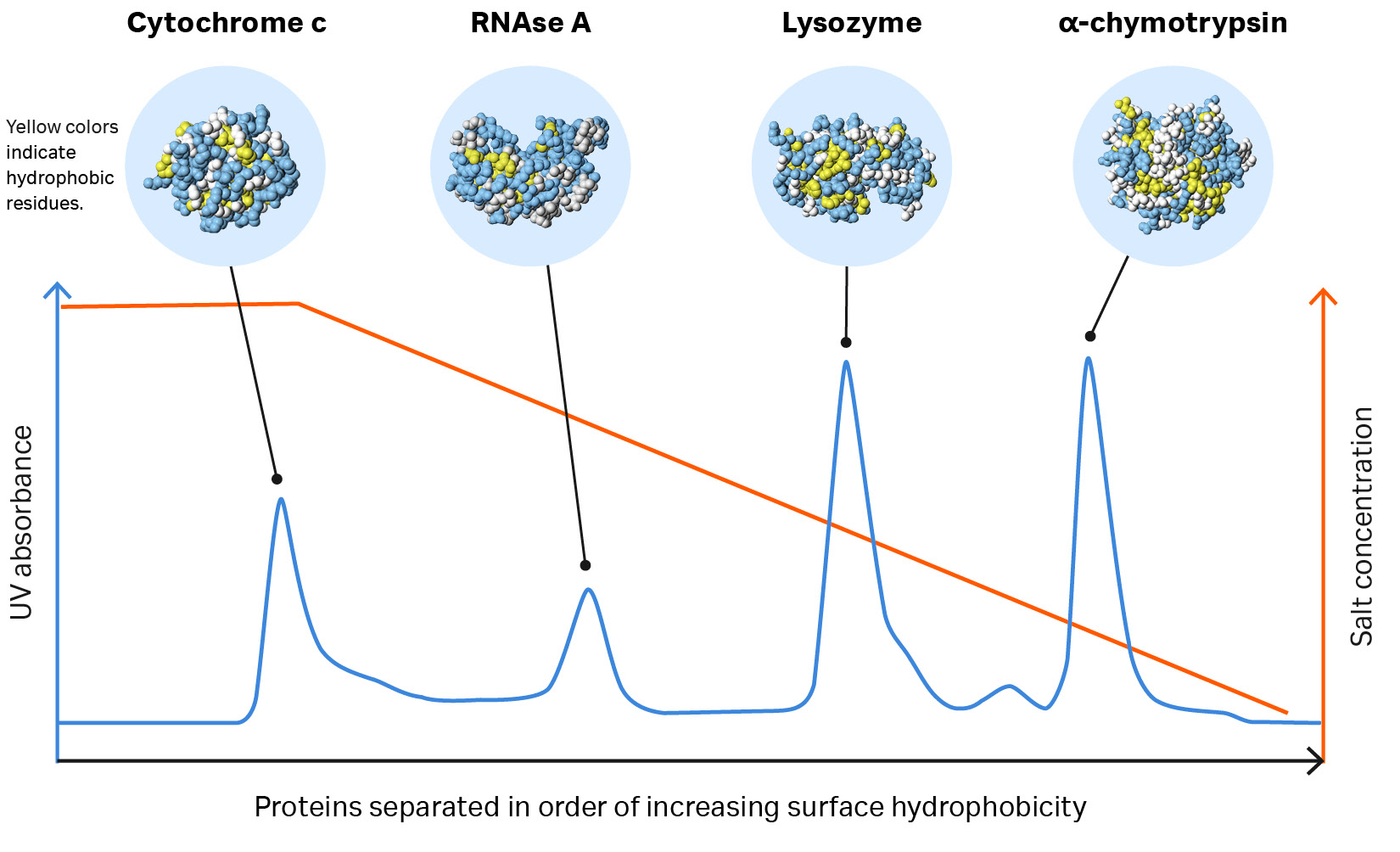
Fig 1. Proteins separate according to differences in their surface hydrophobicity (yellow indicates hydrophobic amino acid residues and blue indicates hydrophilic amino acid residues) as shown in this separation of standard proteins on one of Cytiva’s HIC resins.
You can perform hydrophobic interaction chromatography either in bind/elute, or in flow-through mode. Table 1 shows how you use HIC in four main steps in bind/elute mode. In bind/elute, the target molecule binds to the ligand coupled to the resin through hydrophobic interactions. Changes in the buffer composition and pH release the molecule from the resin to allow collection (the molecule elutes with the buffer).
Table 1. Four main steps of a hydrophobic interaction chromatography that you can perform in bind/elute mode
| Equilibration | Sample application and wash | Elution | Regeneration |
| Prepare the column to the desired start conditions.
Equilibrate HIC resin with high-salt start buffer. |
Target molecule binds to the ligand on the resin via hydrophobic interactions. The sample buffer needs to have the same pH and ionic strength as the starting buffer. |
Decreasing salt content (using a linear gradient) causes hydrophobic proteins to elute. The least hydrophobic proteins elute first. | Removes all molecules still bound to the stationary phase. Ensures the full capacity of the resins is available for the next run. 0.5 to 1.0 M NaOH is often used to clean the resin. |
In flow-through mode, impurities bind to the resin while the target molecule is collected in the chromatography flowthrough.
Watch our animation to easily visualize how hydrophobic chromatography works: