For over 40 yr, immunoassays have been used by hospitals, laboratories, and researchers to rapidly diagnose a wide variety of health, disease, and drug treatment conditions. Information gained by these tests has helped to identify pre-symptomatic disease and improve therapeutic options for patients. In life science research, immunoassays have been used to pinpoint hits, quantitate protein expression levels and sample concentration, and unravel complex molecular interactions to advance biological study, while in industry, they have been used to detect contaminants in our food and water. These quick and accurate tests fall within the broader category of methods frequently described as “binding assays” (Fig 1).
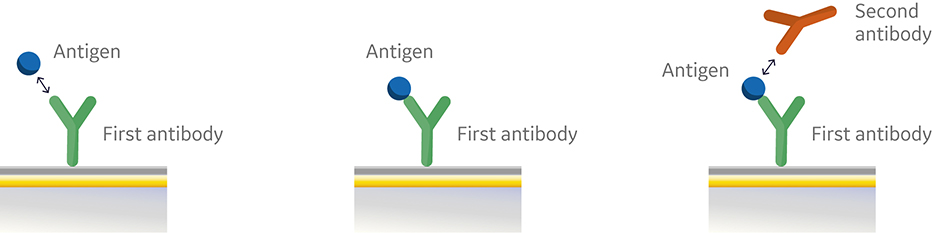
Fig 1. Conventional sandwich immunoassay.
Immunoassay developers are constantly striving to create tests that address unmet diagnostic needs. Doing so would enable them to gain a first-to-market advantage over competitors. But no matter how innovative an assay is, if it does not consistently perform as promised, it is unlikely to generate repeat purchase and will ultimately fail. A critical prerequisite for good assay performance on any technology platform is the design, production, and characterization of high-quality reagents that are highly selective and sensitive.
There are challenges in sourcing reagents that must be addressed in order to develop novel, precise, and fit-for-purpose immunoassays. The following guidelines highlight some critical considerations in developing immunoassay reagents. Steps for avoiding common pitfalls are outlined for reagent developers and for those who develop the assays that depend upon qualified reagents.
Getting started
The key aspects to start the immunoassay development process include selecting the analyte of interest to measure, the sample matrices in which measurements will be made, as well as determining the critical success factors for the assay, for example, sensitivity, throughput, and dynamic range.
This is followed by a screening process, involving antibody reagents, to identify and qualify optimum antibody pairs. Normally, development scientists consider reagents that are already commercially available or internally produced. Then they engage in a lengthy and costly trial-and-error process that may involve testing multiple reagents from a variety of suppliers to find just the right one. According to Cynthia Shuman, PhD, senior applications scientist at Cytiva who works closely with both assay developers and suppliers of reagents, “the key to improve this process, is to start with the desired assay read-out. This will help to remove some of the guesswork in qualifying essential reagents. It’s also useful for reagent suppliers to provide information such as:
- source and identity
- sensitivity as well as specific and selective interactions between reagent and the target analyte
- concentration
- formulation
- storage and use condition
- expiry date.
Armed with this information, assay developers can hone in on the best combination of biological activity and biophysical properties that will yield the desired immunoassay result.
Selection of critical reagents
Suitability
The specificity of reagents towards the analyte of interest is of utmost importance. Binding properties not only determine the applicability of the reagent, but also the sensitivity and robustness of the assay. Assay developers cannot afford to waste time or expense doing experiments with reagents that offer little information regarding primary antibody specificity.
According to Brian Lang, PhD, Biacore sales specialist at Cytiva, “it is important to have a comprehensive biophysical and functional characterization for all reagents, regardless of whether they are obtained from in-house or external suppliers. That means utilizing more information-rich technologies, like surface plasmon resonance,SPR (Fig 2) such as that used in Biacore systems from Cytiva. SPR has the potential to locate epitopes through which reagents interact, as well as very subtle changes in structure (conformational changes, glycosylation, aggregation, etc.) and function.” Having this type of information available helps to identify promising reagents as well as specific sequence modifications that could be made for less antigenic proteins.
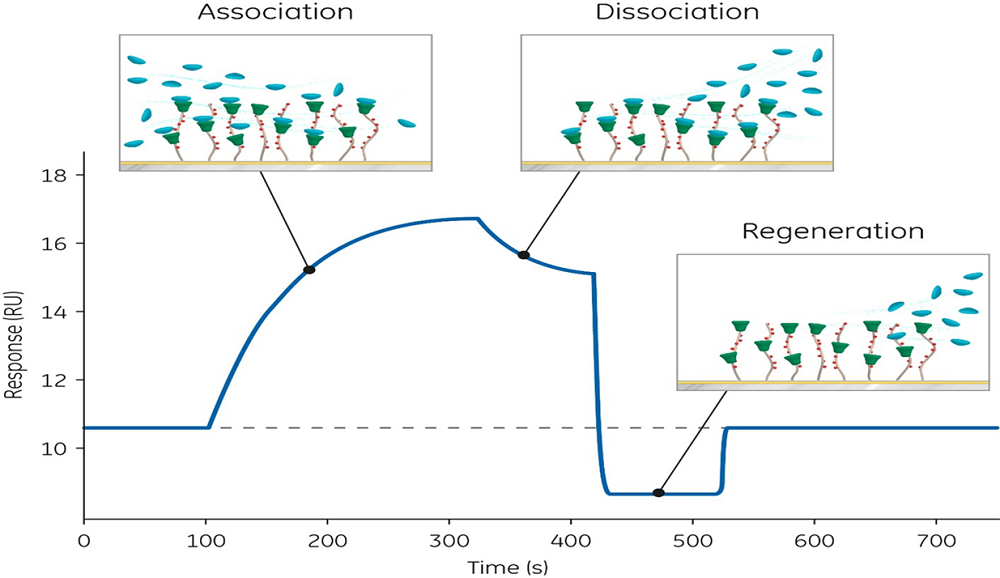
Fig 2. The surface plasmon resonance binding profile, observed in the Biacore sensorgram, provides real-time information about analyte–target binding measured in resonance units (RU).
Binding depends not only on the affinities of the constituent antibodies, but also on their individual concentrations, which are often difficult to ascertain. Elaborates Shuman, “I had an immunoassay developer client, who was working with a reagent that contained too little of the actual active binding antibody concentration. Ultimately, it was difficult to get the assay to work and the development program was canceled, but not before a lot of time and funds were wasted.”
Having the active concentration data available would help to avoid this issue. Suitable analytical methods for reagent concentration assessment include a functional test (e.g., enzyme-linked immunosorbent assay, ELISA or SPR). The SPR method not only provides information on target binding affinity and binding kinetics but also allows for assessment of relative active concentration, and the impact of labeling on reagent properties compared with their unlabeled origins.
Behavior
Binding affinity information alone is not enough when it comes to selecting critical reagents. Reagents that have interacting partners with low molecular weights or are subjected to physicochemical stresses during production, transport or storage (pH, temperature, agitation, and oxidation) can impact binding (affinity, on-rate and off-rate, Fig 3). One reagent may bind faster or slower to the target than another. However, if a particular reagent binds fast, but also disassociates fast, the assay may result in a zero value. Developers really want to quickly identify if there is another antibody with the same affinity that stays bound to the target for longer.
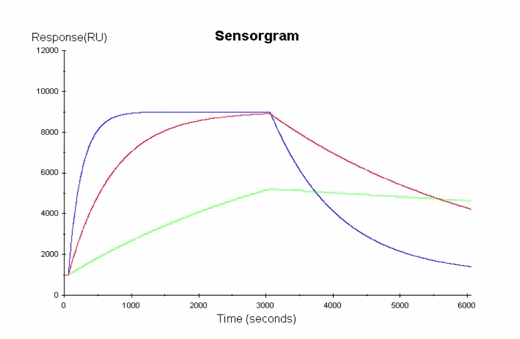
Fig 3. Analyte–target interactions with similar affinities can have very different kinetic properties, which may result in different biological responses. Binding interaction on-rates reflect the recognition between binding partners while off-rates indicate stability of the interaction. By resolving affinity into on- and off-rates, comprehensive information is obtained on how the dynamics of molecular interactions relate to protein function. On-rates reflect recognition between interacting partners while off rates indicate stability of the complex.
It is also important for immunoassay developers to understand whether the reagents are active at a temperature of 37°C (for human samples). If this is not anticipated during original assessments, it might cause a significant setback to assay development. A detailed description of the reagent storage and handling conditions should be made available by reagent suppliers. Formulation information is also critical because some components (e.g., stabilizing proteins) may interfere with the analytical methods used to characterize the reagent. If there are other stabilizing proteins present in the reagent formulation, it would give a string signal itself in the assay and therefore render the results meaningless. Some commercial immunoassay manufacturers do their best to minimize the risk of such interference, by adding non-immune blocking agents to the reaction buffer.
There are many techniques for performing quick screens on reagent binding such as ELISA which is an end-point technique. End-point techniques offer a snapshot view of interactions providing only basic information such as overall binding strength (affinity). The affinity depends on the ratio of on- and off-rates so that equal affinity interactions can have very different kinetic properties, resulting in different binding properties. Surface plasmon resonance (SPR) can provide key binding data in real-time to discriminate these crucial differences, for example, binding kinetics. SPR can also be used to determine the thermodynamics of molecular interactions to aid assay developers in understanding the fundamental driving force for protein-protein interaction.
Another consideration is the potential for cross-reactivity that generates a result outside the expected reaction. Some naturally occurring antigens are a mixture of macromolecules (for example, from pathogens, toxins, or pollen) comprising several epitopes. Contact with a complex antigen such as a virus will stimulate multiple immune responses to the virus' different macromolecules as well as the individual epitopes of each macromolecule. Cross-reactivity can produce false-positive results and is thus a commonly evaluated parameter for validating assay reagents.
Other factors
Critical reagents are typically produced using biological processes and may be prone to variability as different batches are prepared. This can lead to changes in impurity profiles and altered protein structure. Assay developers employ strategies to control between-lot variation by sourcing materials from the same supplier and lot whenever possible. But this is not an ideal long-term solution. To ensure a sustainable high-quality supply, the reagents should be generated based on reviewed manufacturing protocols with defined specifications, be appropriately characterized for quality control, and studied for long-term stability. Reagent suppliers who employ a quality management system that meets the standards of medical use, will certainly enjoy repeat business and long-term partnerships with assay developers.
Conclusion
There is a continued demand for novel, fit-for-purpose immunoassays. Despite the challenges, it’s important for assay developers to streamline the selection and potential modification of critical reagents to get their tests to market quicker. The SPR method provided by, for example, Biacore SPR systems, enables interaction analysis with greater precision in understanding molecular mechanisms and structure-function relationships, paving the way to an increased understanding of biochemical mechanisms. Importantly, it has the potential to simplify immunoassay reagent selection, which helps get these important tests in clinicians’ hands sooner.